Abstract
Background
Brain image genetics provides enormous opportunities for examining the effects of genetic variations on the brain. Many studies have shown that the structure, function, and abnormality (e.g., those related to Alzheimer’s disease) of the brain are heritable. However, which genetic variations contribute to these phenotypic changes is not completely clear. Advances in neuroimaging and genetics have led us to obtain detailed brain anatomy and genome-wide information. These data offer us new opportunities to identify genetic variations such as single nucleotide polymorphisms (SNPs) that affect brain structure. In this paper, we perform a genome-wide variant-based study, and aim to identify top SNPs or SNP sets which have genetic effects with the largest neuroanotomic coverage at both voxel and region-of-interest (ROI) levels. Based on the voxelwise genome-wide association study (GWAS) results, we used the exhaustive search to find the top SNPs or SNP sets that have the largest voxel-based or ROI-based neuroanatomic coverage. For SNP sets with >2 SNPs, we proposed an efficient genetic algorithm to identify top SNP sets that can cover all ROIs or a specific ROI.
Results
We identified an ensemble of top SNPs, SNP-pairs and SNP-sets, whose effects have the largest neuroanatomic coverage. Experimental results on real imaging genetics data show that the proposed genetic algorithm is superior to the exhaustive search in terms of computational time for identifying top SNP-sets.
Conclusions
We proposed and applied an informatics strategy to identify top SNPs, SNP-pairs and SNP-sets that have genetic effects with the largest neuroanatomic coverage. The proposed genetic algorithm offers an efficient solution to accomplish the task, especially for identifying top SNP-sets.
Similar content being viewed by others
Background
With recent technological advances in acquiring multimodal neuroimaging and high-throughput genetics data, massive amounts of multimodal structural and functional imaging data on the human brain as well as genome-wide genetic data from the same set of subjects have been collected. With the availability of these data sets, brain imaging genetics has becoming an emerging research area, which provides enormous opportunities for examining the effects of genetic variations on the brain. Many studies have shown that the structure, function and abnormality (e.g., those related to Alzheimer’s disease) of the brain are heritable. However, which genetic variations contribute to these phenotypic changes are not completely clear.
To bridge this gap, a number of approaches for finding associations between genetic variations and imaging phenotypes arise. A genome-wide association study (GWAS) [1] conducted by Christopher et al., which links genetic variations such as single nucleotide polymorphisms (SNPs) to imaging phenotypes, mainly analyzed the association between SNPs with measures at regions of interest (ROIs). The voxelwise GWAS (vGWAS) was proposed by Stein et al. [2, 3] to generate detailed three-dimensional maps of the SNP effects on the brain, without requiring defining ROIs on the brain. Huang et al. [4] developed a functional genome-wide association study (FGWAS) method to identify sparse signals in an extremely large search space. Compared to GWAS, FGWAS could improve detection capabilities to discover important genetic variations and gene-environment interactions that affect brain structure and function. Vounou et al. [5] proposed another method for simultaneously selecting SNP variants and binding regions assuming that the signals are sparse. This could reduce the number of SNPs and phenotypes tested. Among these methods, the voxelwise GWAS makes it possible to study the SNPs from a more nuanced perspective, and can capture subtle signals that are easily missed by ROI-based methods [6,7,8].
As the number of SNPs increases, the amount of data increases exponentially. In prior studies, the researchers used a variety of methods to detect two marker effects [9]. Günther F, et,al. built models using neural networks to reveal the effects. There is a problem that the estimated weight cannot be explained [10]. The random Forests was used to build accurate decision trees for the effects [11] and the two-stage grouped sure independence screening [12] was used to detect the causal interactions. To detect the effects, other methods had been devised, such as odds ratio [13], Ant Colony Optimization Algorithm [14] and MegaSNPHunter [15]. However, since the effects of n SNPs is more complicated and the data increases rapidly, the research on it is still a problem to be developed.
Although brain imaging genetics has become an emerging and rapidly growing research field [16,17,18,19], the study of genetic effects on neuroanatomic coverage remains to be an underexplored topic. To bridge the gap, in this paper, we perform a genome-wide variant-based study, and aim to identify top SNPs or SNP sets which have genetic effects with the largest neuroanotomic coverage at both voxel and ROI levels. Based on the voxelwise GWAS results, we use the exhaustive search to find the top SNPs or SNP sets that have the largest voxel-based or ROI-based neuroanatomic coverage. For SNP sets with >2 SNPs, we propose an efficient genetic algorithm to identify top SNP sets that can cover all ROIs or a specific ROI.
Results
Results of single marker effects
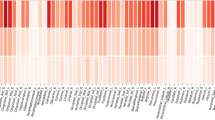
We presented the frequency of the top 10 SNPs with different thresholds from VBC, VBP, RBC and RBP in Fig. 1. Twenty-four SNPs exhibited large neuroanotomic coverage. As expected, the most frequent loci were identified on chromosome 20, including rs6092321 from the RTF2 region, and rs6024860 (N/A). Other SNPs identified in this study are shown in Fig. 1. Table 1 shows the variances explained by identified SNPs. The main effects of rs6092321 and rs6024860 account for 0.93% and 0.82% of phenotypic variance respectively.
The top 10 SNPs and their frequency in VBC, VBP, RBC, RBP. The X axis represents the number of top 10 SNPs in all set thresholds; The Y axis represents the name of SNPs; VBC = Ranking SNPs according to the number of covered voxels; VBP = Ranking SNPs according to minimally required p threshold; RBC = Ranking SNPs according to the number of ROIs; RBP = Ranking SNPs according to minimally required p threshold
Results of SNP–SNP effects
Fifty-three pairs of SNPs showed statistically significant effects on neuroanotomic coverage. Only 1 pair passed the covering criterion: all the four strategy are required to be covered by the SNP pair. The result of SNP–SNP effects was rs6092321 (RTF2) - rs700319 (CNTNAP2). Figure 2 provided the frequency of other SNP pairs. The variance explained by rs6092321 - rs700319 is 0.94%, and the correlation of rs6092321 - rs700319 are 0.0945, 0.0533 and \(-0.0381\) (Table 1).
The top 10 SNP pairs and their frequency in VBC, VBP, RBC, RBP. The X axis represents the number of top 10 SNPs in all set thresholds; The Y axis represents the name of SNP pairs; VBC = Ranking SNPs according to the number of covered voxels; VBP = Ranking SNPs according to minimally required p threshold; RBC = Ranking SNPs according to the number of ROIs; RBP = Ranking SNPs according to minimally required p threshold
Results of three SNPs effects
To find a suitable population size and a maximum evolutionary generation, we chose 100 and 1000 as the center, 0–200 and 0–2000 as the range, and 20 and 200 as the step size to calculate the scores separately. To avoid the occasional final score being too small, we counted the maximum score and the corresponding SNP set after 10 cycles in each case. The resulting score, population size and maximum evolutionary generation are shown in Fig. 3. As shown in Fig. 3, when we used the 100 (population size) and 1000 (maximum evolutionary generation) to filter the SNP sets, the score of the SNP sets reached the highest value of 2.08.
With the selected population size (100) and the maximum evolutionary generation (1000), we counted the relationship between the score and evolutionary generation. The resulting curve is shown in Fig. 4. To avoid the occasional final score being too small or big, we ran the genetic algorithm for 10 times with the same parameters. The scores of the 6th, 7th, 8th, 9th and 10th are around 1.5, and the 6th and 9th converge prematurely. The defects in the initial population and in the offspring may be the major reason. All the remaining scores are above 2.0, and the 1st, 3rd and 4th have the better scores. Their initial scores fluctuate drastically, and the scores increases rapidly over time. Wherein the 1st at about 600th generation converges to optimal score, and the turning point of 3rd and 4th are around 400th generation. This is because the parent with better score that would have an opportunity to breed and pass on their codes.
As expected, the SNP sets with largest neuroanotomic coverage, minimum p value and high number from AR included rs6092321 (Fig. 3). With regard to SR (two ROIs were hippo-campus_L and hippo-campus_R in our experiment), rs429358 was found in most identified SNP sets with the minimum p value (Fig. 5). The results of three SNPs effects were rs6092321 (RTF2) - rs10500192 (CNTNAP2) - rs4811693 (FAM210B) from AR, and rs429358 (APOE) - rs2640726 (EPHX2) - rs4621717 (CNTNAP2), rs429358 (APOE) - rs516125 (SCARA3) - rs4621717 (CNTNAP2), rs429358 (APOE) - rs10933428 (INPP5D) - rs4621717 (CNTNAP2) and rs429358 (APOE) - rs12539907 (CNTNAP2) - rs4621717 (CNTNAP2) from SR. Details are available in Table 1.
The top 10 SNP sets and their frequency in AR and SR. The X axis represents the number of top 10 SNPs in all set thresholds; The Y axis represents the name of SNP sets; AR = Ranking SNPS according to minimal p threshold covered all ROIS; SR = Ranking SNPS according to minimal p threshold covered a specific ROI
Post hoc analysis
Table 1 also shows the correlation between SNPs and hippocampus, memories and memory. For each SNP, SNP pair and SNP set, we superimposed the voxel images of each group of SNPs. We combined the images and features [20] to determine the contribution of SNPs to brain features. For rs6092321, the correlation account for 0.073, 0.0806 and 0.0042, the correlation of rs6092321 - rs700319 account for 0.0945, 0.0533 and \(-0.0381\), and the correlation of rs6092321 - rs10500192 - rs4811693 are 0.0804, 0.0675 and -0.0111. In SR, the correlation of rs429358 - rs12539907 - rs4621717 are 0.3085, 0.1522 and 0.1186.
Discussion
In this work, we performed voxelwise GWAS and using a sample of 1515 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. To our knowledge, this study on detecting SNP, SNP pairs and SNP sets is the first study of genetic effects on neuroanatomic coverage.
The down-sampled image were obtained under the different treatments and used for vGWAS with the genetic data. Then the resulting data analyzed using four computational programs (VBC, VBP, RBC, and RBP) for ranking the SNPs. SNPs were selected based on the number of voxels less than the set p value threshold in VBC, and VBP served as a control group to filter SNPs based on the p value corresponding to the given number of voxels. The primary purpose of VBP is to find the “missing SNPs” in VBC. As illustrated in Fig. 1, the top 9 SNPs are the same and a few SNPs are different in VBC and VBP. The difference of p value in voxels of SNPs accounts for the major cause. Selecting SNPs that affect ROIs and differ with chosen SNPs on voxel level is the main objective in RBC and RBP. The ROI coverage was added to RBC as an additional condition on the basis of VBC. Similar to RBC, the given number of voxels was changed to the set number of ROIs, and the coverage of ROI was added in RBP. Picking SNPs from another condition and comparing them with SNPs in RBC are the primary aim of RBP. A similar discrepancy among SNPs was observed on voxel level and ROI level and directly correlated to the addition of ROI coverage.
According to the four different programs, different rankings of the top SNPs were shown and several missing SNPs were found. These highlight the necessity of utilizing multiple procedures to obtain the best possible SNPs. The frequency of the SNPs can be observed directly by the number, and the lower values indicate “missing SNPs” that have been recovered.
For the number of SNP set selected equal or greater than 3, the number of SNP sets reaches 900 million, and the time of exhaustive strategies increases exponentially. Therefore, algorithms to shorten the time or reduce the data set was developed. Genetic algorithm is suitable to resolve the issue. The initial population of genetic algorithm can be considered a reduced data set, and the offspring after cross-inheritance can be treated as a new data set that is constantly changing. Importantly, when an offspring with a large score appears, the data set will quickly converge to this score, which greatly shortens the selection time. As shown in Fig. 4, the dramatic shifts in scores are about 3.5 and 2.8, and the scores afterwards plateau at this value. And the time for 1000 evolutionary generation can be shortened to less than 20 min.
In single-marker analysis, the most obvious SNP identified from the analysis is rs6092321 (within the RTF2 gene on chromosome 20). The ROIs affected by SNP rs6092321 (RTF2) with a high coverage are left gyrus rectus, right gyrus rectus, left entorhinal cortex, right entorhinal cortex, and vermis_9. The specific function for left gyrus rectus and right gyrus rectus has not yet been brought to light. However, subgroup analysis showed the negative impact of gyrus rectus resection on language and memory recall categories [21]. Ballmaier M. et al. had reported that very significant strong gray matter defects were observed in the gyrus rectus [22]. The volume of the left entorhinal cortex was different between progressive (Alzheimer’s disease) and stable mild cognitive patients [23]. MRIs show that Gomez-Lopez-Hernandez syndrome is characterized by cerebellar sacral loss or partial cerebellar loss and varying degrees of cerebellar fusion [1]. In terms of images, after calculating the Pearson correlation between rs6092321 (RTF2) and features (hippocampus, memories, memory and speaker), we found that rs6092321 (RTF2) is positively correlated with hippocampus, memories, memory and speaker (Additional file 1). This gives an affirmation of the impact of rs6092321 on ROIs above.
In SNP–SNP analysis, rs700319 (within the CNTNAP2 gene on chromosome 7) and rs6092321 (RTF2) appear in pairs with highest frequency, since the frequency of 700319 (CNTNAP2) in single marker analysis was not high. The variances explained by rs6092321 (RTF2) - rs700319 (CNTNAP2) is bigger than rs6092321 alone. Combined with additional file 1, these give the evidence that rs700319 (CNTNAP2) is another important SNP. Comparing with rs6092321 (RTF2), rs6092321 (RTF2) - rs700319 (CNTNAP2) is positively correlated with memories and hippocampus, and negatively correlated with memory (Table 2).
Although the results obtained by the genetic algorithm are not the global optimal solutions, the running time is reduced greatly. And the result achieves improvement with the increase of running time. In AR, considering the variances explained by rs6092321 (RTF2) and rs6092321 (RTF2) - rs10500192 (CNTNAP2) - rs4811693 (FAM210B) and the additional file 1, rs10500192 (CNTNAP2) and rs4811693 (FAM210B) are the “missing SNPs” for the genetic effects on neuroanatomic coverage.
In SR, we found a sudden increase in the number of rs429358 (APOE). A meta-analysis estimated that the ratio of homozygous rs429358 (C;C) individuals to the more common ApoE3 / ApoE3 homozygote was 12 times of late-onset Alzheimer’s disease and 61 times of early-onset disease [24]. These results confirm our prediction and prove that the candidate SNPs we selected will provide more valuable information. For the SNP sets including rs429358 (APOE), the pearson correlation between SNP sets and features (hippocampus, memories and memory) show that the SNP sets have a positive correlation on these features. This suggests that SNP group makes sense for hippocampus, and memory and is also consistent with our initial vision. Considering the difference among the SNP sets including rs429358 (APOE) and the additional file 1, rs12539907 (CNTNAP2) is a “missing SNP” for hippocampus.
Based on the above, the loci including the “missing SNPs” identified in our analysis are CNTNAP2 and FAM210B. CNTNAP2 has an pivotal effect in maintaining normal network activity and synaptic transmission. The transsynaptic bridge formed by CNTNAP2 on the presynaptic membrane and CNTN2 on the post-synaptic membrane can spans the synaptic cleft [25,26,27]. The dendritic arborization and the numbers of excitatory synapses, inhibitory interneurons, and inhibitory synapses were all reduced by the loss of CNTNAP2 [28,29,30]. CNTNAP2 guides the cellular migration of neurons to their correct position in the brain [28, 31]. The impact of CNTNAP2 on cellular migration of neurons, synapse development, and synaptic communication indicate that it plays a key role in the brain function. FAM210B can promote the transfer of protoporphyrinogen IX (PPIX) to FECH, and promote the introduction of iron and the synthesis of heme by forming oligomers with PPOX and FECH to enhance the introduction of mitochondrial iron and the synthesis of heme [32]. Stabilization of FECH protein caused by the binding of iron-sulfur clusters [33] or the increased transcription of FECH mRNA [34] lead to ferrochelatase protein expression increased during erythropoiesis. FAM210B can effectively transport iron to FECH, and / or affect the allosteric activation of the FECH enzyme [32]. The possible mechanisms behind APOE-CNTNAP2 and RTF2-CNTNAP2 warrant further investigation.
In summary, some of the SNPs and genes identified in our analysis have shown interesting associations with the genetic effects on neuroanatomic coverage from prior knowledge of current literatures, such as rs6092321, rs429358, RTF2, APOE and CNTNAP2. The additional file 1 showed the correlation between the brain structure of identified SNPs, SNP pair and SNP sets and the features provide by [20]. These results were very encouraging and confirmed that the analysis was successful as it was able to identify the “missing SNPs” and the top SNPs that have largest neuroanatomic coverage. In addition, numerous SNPs, SNP pairs and SNP sets revealed in our study had genetic effects, which warrant further investigation or replication in future studies.
The limitations of our study are as follows: (1) We examined 1784 SNPs. We also need use more SNPs to analyze. (2) We used the genetic algorithm to analyze the effects of multiple SNPs. The result is a local optimal solution and more effective and efficient strategies are still to be developed in multiple SNPs. (3) Comparing the exhaustive search, we can get better results in less time using genetic algorithms. However, the results get better with time, and users still have to wait a long time to get better results. (4) When a better set of SNPs appears, the offspring will be stuck in this combination.
Conclusion
Aiming at studying the relationship between SNPs and brain structures, we performed voxel-wise GWAS and SNPs analysis to discover the SNPs which could affect more areas of the brain based on ROI and voxel using a sample of 1515 subjects from the ADNI database. The single-marker analysis identified the SNPs rs6092321 and rs6024860, which contributed the highest genetic effects on neuroanatomic coverage in all case. The SNP–SNP analysis identified new SNP pair including rs6092321 in single-marker analysis, which showed strong associations with the neuroanatomic coverage. This was rs6092321 and rs700319. The n SNPs analysis identified a number of novel findings, which showed higher associations with whole brain or hippocampus. Perhaps what is more important in this study is the discovery of SNPs that has not yet been associated with the Alzheimer’s Disease (AD) in conventional GWAS studies. Based on voxelwise GWAS, the effects of n SNPs and SNP–SNP showed high-level statistical significance than the single-marker effects. These may help address part of missing SNPs and brain clusters. Although this study focuses on SNPs effects, the findings may well show that the genetic algorithm is an interesting method for detecting the effects of n SNPs.
Our voxelwise genome-wide association study and genetic effects study on neuroanatomic coverage have the following strengths in addition to the above interesting findings. (1) To our knowledge this is the first study of genetic effects on neuroanatomic coverage. (2) Using voxelwise volumetric measurements as phenotypes confers higher statistical power than using conventional phenotypes and is able to find the “missing SNPs”. (3) The sample in this study included HC, SMCI, EMCI, LMCI, and AD, thus providing a rank-ordered spectrum of the disease progression. (4) Our approach is more computationally efficient than the exhaustive strategies, facilitating the analysis of genome-wide SNPs effects.
Methods
We first describe the imaging and genotype data used in this work and then present our methods.
Imaging and Genotype Data
The imaging and genotype data of 1,515 non-Hispanic Caucasian subjects were downloaded from http://adni.loni.usc.edu. In this work, we analyzed the MRI scans and genoty** data, including 353 healthy control (HC), 89 significant memory concern (SMC), 273 early MCI (EMCI), 504 late MCI (LMCI), and 296 AD participants. The characteristics of these 1,515 subjects are shown in Table 2.
Preprocessed T1-weighted volumetric MRI scans were aligned to each participant’s same visit scan and normalized to the Montreal Neurological Institute (MNI) space. Voxel-based morphometry (VBM) was applied on MRI scans to extract voxel-wise volumetric measurements. Briefly, scans were aligned to a T1-weighted template image, segmented into gray matter, white matter and cerebrospinal fluid maps, and then normalized to the MNI space. The gray matter density (GMD) maps were extracted and smoothed with an 8mm FWHM kernel. The resulting GMD images are then down-sampled to a dimension of \(61 \times 73 \times 61\) (i.e., containing 271,633 voxels) to reduce the computation cost in subsequent analyses. The 116 ROIs and their coordinates were anatomically defined using the Automatic Anatomical Labeling (AAL) atlas [35], and registered with the down-sampled images.
Genoty** data was processed as described in [36, 37], which resulted in 565,373 SNPs for all 1515 participants. A list of 24 AD candidate genes from a prior large scale GWAS meta-analysis [38] were analyzed in this study. We extracted SNPs located in \(\pm 20\)K bp of the 24 AD genes, and finally included total 1784 SNPs in our imaging genetic association analysis.
Overall strategy
As shown in Fig. 4, the first step of our investigation is to perform pairwise univariate voxelwise genetic association analysis on 1515 subjects to examine the variant effect of 1784 SNPs on 271,633 voxels of the brain. The p value of each SNP-voxel pair was first obtained by performing genetic association of all the voxels for each studied SNP. Using these voxelwise SNP results, we implemented the following four strategies to identify top SNPs or SNP pairs that affect the largest portion of the neuroanatomy on the voxel or ROI basis.
-
1
VBC: Given a p threshold, rank SNPs according to the number of significant voxels.
-
2
VBP: Given a voxel number threshold, rank SNPs according to minimally required p threshold.
-
3
RBC: Given a p threshold and ROI coverage threshold, rank SNPs according to the number of ROIs covered by an enough number of significant voxels.
-
4
RBP: Given an ROI number threshold and ROI coverage threshold, rank SNPs according to minimally required p threshold.
Although the exhaustive search strategy can be applied to identify top SNPs or SNP pairs, it won’t work on identifying SNP sets containing three or more SNPs (denoted as high-order SNP sets for convenience) due to exponentially increasing computational cost. To address this challenge, we propose a more efficient genetic algorithm to identify top high-order SNP-sets, whose effects have the largest neuroanatomic coverage.
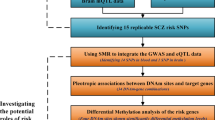
The schematic design of the workflow. VBC = Ranking SNPs according to the number of covered voxels; VBP = Ranking SNPs according to minimally required p threshold; RBC = Ranking SNPs according to the number of ROIs; RBP = Ranking SNPs according to minimally required p threshold; AR = Ranking SNPS according to minimal p threshold covered all ROIS; SR = Ranking SNPS according to minimal p threshold covered a specific ROI; top SNPs, SNP pairs and SNP sets = the identified top SNPs, SNP pairs and SNP sets which have genetic effects with the largest neuroanotomic coverage or specific ROI
Figure 6 shows a schematic design of the workflow of our analyses. In the following subsections, we describe these analyses in more detail.
Identification and prioritization of single marker effects
VBC: ranking SNPs according to the number of covered voxels. To get the number of voxels, we defined the score for each SNP as the number of voxels who had a p value smaller than a threshold. Then we got a list of SNPs sorted by the number of covered voxels in descending order (Fig. 7). To avoid accidental SNPs, we set multiple thresholds. The top 10 frequent SNPs of the results were shown in Fig. 1.
VBP: ranking SNPs according to minimal p value threshold. For finding the minimal p value of each SNP in this criterion, we set a condition that the number of voxels was limited (Fig. 8). Figure 1 showed the top 10 SNPs of the results limited by a number of thresholds.
To determine the union of voxels representing the ROI, one way was to calculate the weighted average of the ROI and the other was to select voxels above a certain threshold [39]. In our study, we used the percentile (at least 20%) and p value (\(p < 0.05\)) of voxels as the threshold.
RBC: ranking SNPs according to number of covered ROIs. Like the VBC, to get the number of covered ROIs, we defined how one SNP affected a ROI. In a similar vein, given a threshold on p value, one SNP was considered affecting a ROI if it covered 20% voxels of this ROI. Each SNP was ranked based on the number of the ROIs that it affected (Fig. 9). Figure 1 presented the top 10 SNPs.
RBP: ranking SNPs according to minimal p value threshold. The goal of this section was to got a minimal p value based on ROI. Each SNP was ranked according to the minimal p value covered a given number of ROIs. In our experiments, in the condition of covering over 20% voxels of the ROI and the number of the ROI was set, one SNP could be taken into the selection (Fig. 10). The top 10 SNPs with the highest frequent were presented in Fig. 1.
Identification and prioritization of SNP–SNP effects
For SNP–SNP, a new concept was imported to determine the p value within the effects of SNP–SNP. To acquire an operable p value of SNP–SNP, we took the minimal p value of SNP–SNP on one voxel as the upshot since we used single marker effects. The main strategy of SNP–SNP effects were similar to VBC, VBP, RBC and RBP.
The schematic diagram of ranking SNPs according to number of covered ROIs. 1. Calculate the percentage of voxels with p value less than the set threshold in each ROI; 2. Rank ROIs according to the percentages; 3. Calculate the number of ROIs with voxels’ percentage more than the set value in each SNP; 4. Rank SNPs according to the numbers
The schematic diagram of ranking SNPs According to minimal p value threshold. 1. Rank voxels according to p value in each ROI; 2. Calculate the p value on the set voxel node; 3. Rank ROIs according to the p value; 4. Calculate the p value on the set ROI node in each SNP; 5. Rank SNPs according to the p value
Identification and prioritization of three SNP effects
AR: ranking SNPs according to minimal p value covered all ROIs. In the effects of the three SNPs, the exhaustive search could take more than 30 days. Therefore, we improved the genetic algorithm to make it more suitable for our experiments. There were four main steps in genetic algorithms (Fig. 11).
The workflow of the genetic algorithms. Coding = the SNP sets code into a 3-bit code; Initialization = the generation of initial data set; Tournament Strategy = mining the best code from a set of codes; Intersection = exchanging parents’ code in a random site; Mutation = generating a new 3-bit code; Offspring = the result of Intersection or Mutation; Fitness function = based on the p value of 3 SNPs affecting 116 ROIs or a specific ROI, the SNPs sets with minimal p value are picked out as the parent; Decoding = the 3-bit code decode into corresponding SNP sets
Step 1, coding and initialization.
Step 2, fitness function and selection.
Step 3, Intersection and mutation.
Step 4, decoding.
In order to facilitate the analysis, the minimal p value of the three SNPs was set on one voxel as the consequence under the same experimental criterion of the two SNPs.
In step 1, considering the set of p value on 271,633 voxels of 1784 SNPs, the coding strategy and decoding strategy were established. Since it was the effects of three SNPs, the coding strategy was determined to be a 3-bit code, and each bit was 1 to 1784.
In step 2, to filter out the parent, tournament strategy was introduced in this section. For choosing the parent in the tournament strategy, we defined the fitness function: based on the p value of 3 SNPs affecting 116 ROIs, the SNPs sets with larger score were picked out as the parent. The score was defined in formula 1. We first considered the p value of SNPs, and then calculated the coverage of SNPs in ROIs. Therefore, when \(p value > 0.05\), the proportion of p value was 0. When \(p = 0.05\), the proportion of coverage and p value were same. When \(p < 0.05\), the proportion of p value was greater than the coverage.
where p is the maximum pvalue of 3 SNPs. cov is the maximum coverage of 116 ROIs. \({\log _{0.05}0.2}\) is used to modify the score of coverage \(= 20\%\) to \({-\log _{10}0.05}\).
In step 3, for yielding progeny populations, a random parameter called the probability of intersection was defined. If it was less than \(P_c\), a random position was chosen as the intersection from the 3-bit code for crossover operation. To avoid data locking in a combination, another parameter called probability of variation was generated randomly and compared with \(P_m\). If it was less than \(P_m\), a mutation operation would be performed.
The adaptive values of Pc and Pm were determined using formula 2 and 3 [40].
where \(k_1, k_2, k_3, k_4 \le 1.0\). \(f_{max}\) is the maximum score of the population. \(f^{'}\) is the larger score of two intersecting individuals. \({\overline{f}}\) is the average score of the population. f is the score of offspring.
In step 4, the results were decoded into corresponding SNP sets.
SR: ranking SNPs according to minimal p value covered a specific ROIs. The fitness function in step 2 was defined as: based on the p value of 3 SNPs affecting a sprcific ROI (the coverage > 20% and the biggest coverage of other ROIs < 15%), the SNPs sets with minimal p value were picked out as the parent. Other strategy of SR were similar to AR.
Availability of data and materials
Data used in this article was downloaded from the ADNI database (http://adni.loni.usc.edu). Application for access to the ADNI data can be submitted by anyone at http://adni.loni.usc.edu/data-samples/access-data/.
Abbreviations
- AAL:
-
Automatic anatomical labeling
- AD:
-
The Alzheimer’s disease
- ADNI:
-
The Alzheimer’s disease neuroimaging initiative
- AR:
-
All ROIs
- EMCI:
-
Early mild cognitive complaint
- FGWAS:
-
Functional genome-wide association study
- GMD:
-
The gray matter density
- GWAS:
-
Voxelwise genome-wide association study
- HC:
-
Healthy control
- LMCI:
-
Late mild cognitive complaint
- MCI:
-
Mild cognitive impairment
- MNI:
-
Montreal neurological institute
- MRI:
-
Magnetic resonance imaging
- PET:
-
Positron emission tomography
- PPIX:
-
Protoporphyrinogen IX
- RBC:
-
ROI-based coverage
- RBP:
-
ROI-based p value
- ROI:
-
Region-of-interest
- SMC:
-
Significant memory concern
- SNPs:
-
Single nucleotide polymorphisms
- SR:
-
The specific ROIs
- VBC:
-
Voxel-based coverage
- VBM:
-
Voxel-based morphometry
- VBP:
-
Voxel-based p value
- vGWAS:
-
Voxelwise GWAS
References
Newton-Cheh C, Hirschhorn JN. Genetic association studies of complex traits: design and analysis issues 573(1–2):54–69
Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, Saykin AJ, Shen L, Foroud T, Pankratz N. Voxelwise genome-wide association study (vgwas). Neuroimage 53(3):1160–1174
Hibar DP, Stein JL, Kohannim O, Jahanshad N, Saykin AJ, Shen L, Kim S, Pankratz N, Foroud T, Huentelman MJ. Voxelwise gene-wide association study (vgenewas): Multivariate gene-based association testing in 731 elderly subjects. Neuroimage 56(4):1875–1891
Huang C, Thompson P, Wang Y, Yu Y, Zhu H. Fgwas: Functional genome wide association analysis. Neuroimage; 2017. p. 159.
Vounou M, Nichols TE, Montana G. Discovering genetic associations with high-dimensional neuroimaging phenotypes: A sparse reduced-rank regression approach. Neuroimage 53(3):1147–1159
Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW. Common alzheimer’s disease risk variant within the clu gene affects white matter microstructure in young adults. Journal of Neuroscience 31(18):6764–6770
Hibar D, Stein J, Jahanshad N, Kohannim O, Hua X, Toga A, McMahon K, de Zubicaray G, Martin N, Wright M, Weiner M, Thompson P. Genome-wide interaction analysis reveals replicated epistatic effects on brain structure. Neurobiol Aging. 2015;36:151–8.
Liu J, Calhoun V. A review of multivariate analyses in imaging genetics. Front Neuroinform. 2014;8:29.
Koo CL, Liew MJ, Mohamad MS, Mohamed Salleh AH. A review for detecting gene–gene interactions using machine learning methods in genetic epidemiology. BioMed Res Int. 2013;2013:1–13. https://doi.org/10.1155/2013/432375.
Günther F, Wawro N, Bammann K. Neural networks for modeling gene–gene interactions in association studies. BMC Genet. 2009;10(1):87. https://doi.org/10.1186/1471-2156-10-87.
Nguyen T, Le L. Detection of SNP-SNP Interactions in Genome-wide Association Data Using Random Forests and Association Rules. In: 2018 12TH INTERNATIONAL CONFERENCE ON SOFTWARE, KNOWLEDGE, INFORMATION MANAGEMENT & APPLICATIONS (SKIMA), pp. 1–7 (2018). IEEE Islamabad Sect; Glink; Smart Link; Leader
Fang Y-H, Wang J-H, Hsiung CA. Tsgsis: a high-dimensional grouped variable selection approach for detection of whole-genome snp–snp interactions. Bioinformatics. 2017;33(22):3595–602. https://doi.org/10.1093/bioinformatics/btx409.
Wu X, Dong H, Luo L, Zhu Y, Peng G, Reveille JD, **ong M. A novel statistic for genome-wide interaction analysis. PLoS Genet. 2010;6(9):1001131. https://doi.org/10.1371/journal.pgen.1001131.
Sun Y, Shang J, Liu JX, Li S. An improved ant colony optimization algorithm for the detection of snp–snp interactions. In: Intelligent Computing Methodologies, vol. 9773 (2016). Springer
Wan X, Yang C, Yang Q, Xue H, Tang NL, Yu W. Megasnphunter: a learning approach to detect disease predisposition snps and high level interactions in genome wide association study. BMC Bioinform. 2009;10(1):13. https://doi.org/10.1186/1471-2105-10-13.
Shen L, Thompson PM. Brain imaging genomics: integrated analysis and machine learning. Proc IEEE. 2020;108(1):125–62.
Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814.
...Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, Green RC, Hu X, Huentelman MJ, Kim S, Kauwe JS, Li Q, Liu E, Macciardi F, Moore JH, Munsie L, Nho K, Ramanan VK, Risacher SL, Stone DJ, Swaminathan S, Toga AW, Weiner MW, Saykin AJ. Alzheimer’s Disease Neuroimaging I. Genetic analysis of quantitative phenotypes in ad and mci: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207.
Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, Foroud T, Pankratz N, Moore JH, Sloan CD, Huentelman MJ, Craig DW, Dechairo BM, Potkin SG, Jack JCR, Weiner MW, Saykin AJ. Alzheimer’s Disease Neuroimaging, I: whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in mci and ad: A study of the adni cohort. Neuroimage. 2010;53(3):1051–63.
Wager T. Neurosynth: a new platform for large-scale automated synthesis of human functional neuroimaging data. Front Neuroinform. 2011;5(6):799–801.
Joo MS, Park DS, Moon CT, Chun YI, Song SW, Roh HG. Relationship between gyrus rectus resection and cognitive impairment after surgery for ruptured anterior communicating artery aneurysms. J Cerebrovasc Endovasc Neurosurg. 2016;18(3):223–8.
Ballmaier M, Toga AW, Blanton RE, Sowell ER, Lavretsky H, Peterson J, Pham D, Kumar A. Anterior cingulate, gyrus rectus, and orbitofrontal abnormalities in elderly depressed patients: An mri-based parcellation of the prefrontal cortex. Am J Psychiatry 161(1):99–108
Lopez ME, Bruna R, Aurtenetxe S, Pineda-Pardo JA, Marcos A, Arrazola J, Reinoso AI, Montejo P, Bajo R, Maestu F. Alpha-band hypersynchronization in progressive mild cognitive impairment: a magnetoencephalography study. J Neurosci Off J Soc Neurosci 34(44):14551–14559
Rubinsztein DC, Easton DF. Apolipoprotein e genetic variation and alzheimer & rsquos disease. Dementia Geriatric Cogn Disorders. 1999;10(3):199–209.
Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am J Human Genet 2008;82(1):165–173
Pinatel D, Hivert B, Boucraut J, Saint Martin M, Rogemond V, Zoupi L, Karagogeos D, Honnorat J, Faivre-Sarrailh C. Inhibitory axons are targeted in hippocampal cell culture by anti-caspr2 autoantibodies associated with limbic encephalitis. Front Cell Neurosci. 2015;9:265.
Varea O, Martin-De-Saavedra MD, Kopeikina KJ, Schürmann B, Fleming HJ, Fawcett-Patel JM, Bach A, Jang S, Peles E, Kim E. Synaptic abnormalities and cytoplasmic glutamate receptor aggregates in contactin associated protein-like 2/caspr 2 knockout neurons. Proc Nat Acad Sci USA. 2015;112(19):6176–81.
Peñagarikano O, Abrahams B, Herman E, Winden K, Gdalyahu A, Dong H, Sonnenblick L, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg J, Peles E, Geschwind D. Absence of cntnap2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–46.
Anderson GR, Galfin T, Xu W, Aoto J, Malenka RC, Sudhof TC. Candidate autism gene screen identifies critical role for cell-adhesion molecule caspr2 in dendritic arborization and spine development. Proc Nat Acad Sci. 2012;109(44):18120–5.
Amos G, Maria L, Olga P, Peyman G, Trachtenberg JT, Gescwind DH, Anna D. The autism related protein contactin-associated protein-like 2 (cntnap2) stabilizes new spines: an in vivo mouse study. PLoS ONE. 2015;10(5):0125633.
Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2 - nejm. Digest World Core Med J. 2006;354(13):1370–7.
Yien YY, Shi J, Chen C, Cheung JTM, Grillo AS, Shrestha R, Li L, Zhang X, Kafina MD, Kingsley PD, et al. Fam210b is an erythropoietin target and regulates erythroid heme synthesis by controlling mitochondrial iron import and ferrochelatase activity. J Biol Chem. 2018;293(51):19797–811.
Ning B, Liu G, Liu Y, Su X, Anderson GJ, Zheng X, Chang Y, Guo M, Liu Y, Zhao Y, et al. 5-aza-2’-deoxycytidine activates iron uptake and heme biosynthesis by increasing c-myc nuclear localization and binding to the e-boxes of transferrin receptor 1 (tfr1) and ferrochelatase (fech) genes. J Biol Chem. 2011;286(43):37196–206.
Crooks DR, Ghosh MC, Haller RG, Tong W-H, Rouault TA. Posttranslational stability of the heme biosynthetic enzyme ferrochelatase is dependent on iron availability and intact iron-sulfur cluster assembly machinery. Blood. 2010;115(4):860–9.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M. Automated anatomical labeling of activations in spm using a macroscopic anatomical parcellation of the mni mri single-subject brain. Neuroimage. 2002;15(1):273–89.
Yao X, Cong S, Yan J, Risacher SL, Saykin AJ, Moore JH, Shen L. Regional imaging genetic enrichment analysis. Bioinformatics. 2019.
Yao X, et al. Targeted genetic analysis of cerebral blood flow imaging phenotypes implicates the INPP5D gene. Neurobiol Aging. 2019;81:213–21.
Lambert JC, Ibrahimverbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer’s disease 2013;9(4):1452–8
Mitsis GD, Iannetti GD, Smart TS, Tracey I, Wise RG. Regions of interest analysis in pharmacological fmri: How do the definition criteria influence the inferred result? Neuroimage 40(1):121–132
Srinivas M, Patnaik LM. Adaptive probabilities of crossover and mutation in genetic algorithms. IEEE Trans Syst Man Cybern. 1994;24(4):656–67. https://doi.org/10.1109/21.286385.
Acknowledgements
The complete ADNI Acknowledgement is available at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Funding
The project was partially supported by the China Scholarship Fund (201806680080) and the National Natural Science Foundation of China (61773134 and 61803117), the Natural Science Foundation of Heilongjiang Province of China (YQ2019F003), the Fundamental Research Funds for the Central Universities (3072020CF0402) at Harbin Engineering University; and by Natural Science Foundation of Heilongjiang Province of China (QC2018080), MOE (Ministry of Education in China) Project of Humanities and Social Sciences (19YJCZH120), National Natural Science Foundation (NNSF) of China (61901063). Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Contributions
J.L., S.F. and H.L. led and supervised the research. J.L., W.L., H.L., S.F. and H.L. designed the research and wrote the article. W.L. performed the data processing, visualization of results, system development, SNP analysis and gene analysis. F.C., H.L., P.B., Y.L., H.J. and Y.G. provided suggestions for separation and matching of brain images. S.F. and H.L. provided guidance and consultation on the genoty** and biomarker details about ADNI data, data preprocessing, quality control, GWAS protocol and gene analysis. All the authors reviewed, commented and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study procedures were approved by the institutional review boards of all participating centers (http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf), and written informed consent was obtained from all participants or their authorized representatives. Ethics approval was obtained from the institutional review boards of each institution involved: Oregon Health and Science University; University of Southern California; University of California-San Diego; University of Michigan; Mayo Clinic, Rochester; Baylor College of Medicine; Columbia University Medical Center; Washington University, St. Louis; University of Alabama at Birmingham; Mount Sinai School of Medicine; Rush University Medical Center; Wien Center; Johns Hopkins University; New York University; Duke University Medical Center; University of Pennsylvania; University of Kentucky; University of Pittsburgh; University of Rochester Medical Center; University of California, Irvine; University of Texas Southwestern Medical School; Emory University; University of Kansas, Medical Center; University of California, Los Angeles; Mayo Clinic, Jacksonville; Indiana University; Yale University School of Medicine; McGill University, Montreal-Jewish General Hospital; Sunnybrook Health Sciences, Ontario; U.B.C.Clinic for AD & Related Disorders; Cognitive Neurology-St. Joseph’s, Ontario; Cleveland Clinic Lou Ruvo Center for Brain Health; Northwestern University; Premiere Research Inst (Palm Beach Neurology); Georgetown University Medical Center; Brigham and Women’s Hospital; Stanford University; Banner Sun Health Research Institute; Boston University; Howard University; Case Western Reserve University; University of California, Davis-Sacramento; Neurological Care of CNY; Parkwood Hospital; University of Wisconsin; University of California, Irvine-BIC; Banner Alzheimer’s Institute; Dent Neurologic Institute; Ohio State University; Albany Medical College; Hartford Hospital, Olin Neuropsychiatry Research Center; Dartmouth-Hitchcock Medical Center; Wake Forest University Health Sciences; Rhode Island Hospital; Butler Hospital; UC San Francisco; Medical University South Carolina; St. Joseph’s Health Care Nathan Kline Institute; University of Iowa College of Medicine; Cornell University; and University of South Florida: USF Health Byrd Alzheimer’s Institute.
Consent for publication
Not applicable.
Competing interests
The authors have no actual or potential conflicts of interest including any financial, personal, or other relationships with other people or organizations that could inappropriately influence (bias) our work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
The pearson correlation of SNPs, SNP pair or SNP sets and 3168 features.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, J., Liu, W., Li, H. et al. Genome-wide variant-based study of genetic effects with the largest neuroanatomic coverage. BMC Bioinformatics 22, 223 (2021). https://doi.org/10.1186/s12859-021-04145-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-021-04145-0