Abstract
Background
Gonadotropin releasing hormone (GNRH1) triggers the release of follicle stimulating hormone and luteinizing hormone from the pituitary. Genetic variants in the gene encoding GNRH1 or its receptor may influence breast cancer risk by modulating production of ovarian steroid hormones. We studied the association between breast cancer risk and polymorphisms in genes that code for GNRH1 and its receptor (GNRHR) in the large National Cancer Institute Breast and Prostate Cancer Cohort Consortium (NCI-BPC3).
Methods
We sequenced exons of GNRH1 and GNRHR in 95 invasive breast cancer cases. Resulting single nucleotide polymorphisms (SNPs) were genotyped and used to identify haplotype-tagging SNPs (htSNPS) in a panel of 349 healthy women. The htSNPs were genotyped in 5,603 invasive breast cancer cases and 7,480 controls from the Cancer Prevention Study-II (CPS-II), European Prospective Investigation on Cancer and Nutrition (EPIC), Multiethnic Cohort (MEC), Nurses' Health Study (NHS), and Women's Health Study (WHS). Circulating levels of sex steroids (androstenedione, estradiol, estrone and testosterone) were also measured in 4713 study subjects.
Results
Breast cancer risk was not associated with any polymorphism or haplotype in the GNRH1 and GNRHR genes, nor were there any statistically significant interactions with known breast cancer risk factors. Polymorphisms in these two genes were not strongly associated with circulating hormone levels.
Conclusion
Common variants of the GNRH1 and GNRHR genes are not associated with risk of invasive breast cancer in Caucasians.
Similar content being viewed by others
Background
Exposure to steroid hormones (estrogens and androgens) is a risk factor for breast cancer. Increased exposure to estrogens, for instance by early menarche, late menopause, low parity and post-menopausal obesity, contributes to increased breast cancer risk (reviewed in ref. [1]). High circulating levels of estrogens are associated with elevated breast cancer risk [2, 3].
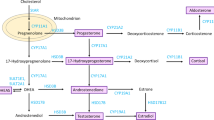
The primary stimulus for production of estrogen and other ovarian steroid hormones is the release of the gonadotropins, follicle-stimulating hormone (FSH) and luteinizing hormone (LH), from the anterior pituitary. These are released when gonadotropin-releasing hormone 1 (GNRH1), from the hypothalamus, binds to the gonadotropin-releasing hormone receptor (GNRHR) in the anterior pituitary. The resultant G-protein activation of a phosphatidylinositol-calcium second messenger system ultimately triggers the release of FSH and LH.
GNRH1 activity is low during childhood but increases at puberty. GNRH1 production is pulsatile. In females, the GNRH1 pulse frequency varies during the menstrual cycle, with a large surge of GNRH1 just before ovulation. The size and frequency of the GNRH1 pulse, and feedback from androgens and estrogens, control production of LH and FSH [4].
GNRH1 activity can be disrupted by hypothalamic-pituitary disease. Elevated prolactin levels decrease GNRH1 activity. In contrast, hyperinsulinemia increases pulse activity leading to disorderly LH and FSH activity, as seen in polycystic ovary syndrome [5].
The GNRH1 gene is located on chromosome 8p21.2. It spans about 5 kb and contains 3 exons. It encodes the GNRH1 precursor, which contains 92 amino acids and is processed to GNRH1, a decapeptide. The GNRHR gene is located on chromosome 4q13.2. Its genomic sequence covers about 19 kb and it includes 3 exons.
The GNRH1 and GNRHR genes can harbor rare germline mutations which lead to idiopathic hypogonadotropic hypogonadism (IHH) or Kallmann syndrome (MIM 146110, 147950) in both men and women [6]. Common variants have not been studied for either gene in relation to cancer risk.
We hypothesized that common, functional polymorphisms of GNRH1 and GNRHR could influence breast cancer risk by modifying production of FSH/LH and steroid hormones. We used a haplotype tagging approach to examine this hypothesis using cases and controls from the BPC3.
Methods
Study Population
The BPC3 has been described in detail elsewhere [7]. Briefly, the consortium includes large, well-established cohorts assembled in the United States and Europe, that have both DNA samples and extensive questionnaire information. These include: the American Cancer Society Cancer Prevention Study II (CPS-II) [8], the European Prospective Investigation into Cancer and Nutrition (EPIC) [9], the Harvard Nurse's Health Study (NHS) [10] and Women's Health Study (WHS) [11], and the Multiethnic Cohort (MEC) [12].
Cases were identified in each cohort by self report with subsequent confirmation of the diagnosis from medical records or tumor registries, and/or linkage with population-based tumor registries (method of confirmation varied by cohort). Controls were matched to cases by ethnicity and age, and in some cohorts, additional criteria, such as country of residence in EPIC.
Most of the subjects were Caucasians of European descent. One cohort (MEC) provided most of the non-Caucasian samples. In total, we genotyped 4,401 Caucasian cases and 5,966 controls, 329 Latino cases and 385 controls, 341 African American cases and 426 controls, 425 Japanese American cases and 418 controls, and 107 Native Hawaiian cases and 285 controls.
Written informed consent was obtained from all subjects, and the project has been approved by the competent institutional review boards for each cohort.
Selection of haplotype tagging single nucleotide polymorphisms (htSNPs)
We sequenced exons and intron/exon junctions of GNRH1 and GNRHR in a panel of 95 metastatic breast cancer cases from the MEC and EPIC. These included 19 cases from each ethnic group represented in the study (African American, Latino, Japanese, Native Hawaiian, and Caucasian). About 45 kb were surveyed for GNRH1 and about 56 kb for GNRHR. No non-synonymous or splice-site variants were identified in sequencing of the exons.
Based on the resequencing and SNPs available in dbSNP, we identified 17 SNPs in GNRH1 and 36 SNPs in GNRHR with minor allele frequency greater than 5% in any of the five ethnic groups or greater than 1% overall. These SNPs were genotyped in a reference panel of 349 healthy women (70 African-Americans, 68 Latinos, 72 Japanese, 70 Caucasians, and 69 Hawaiians from the MEC cohort who had not been diagnosed with breast cancer at the time of the study; average age 65.1 (standard deviation 8.5)) at the Broad Institute (Cambridge, MA, USA) using the Sequenom (San Diego, CA, USA) and Illumina (San Diego, CA, USA) platforms.
Haplotype tagging SNPs (htSNPs) were then selected using the method of Stram et al. [13] to maximize R2 H among Caucasians. Three htSNPs were selected for GNRH1 (including one localized in the 5' neighboring gene, KCTD9, and one in the gene at the 3', DOCK5) and seven for GNRHR.
Genoty**
Genoty** of htSNPs was performed in 3 laboratories (University of Southern California, Los Angeles, CA, USA; Harvard School of Public Health, Boston, MA, USA; International Agency for Research on Cancer, Lyon, France) using a fluorescent 5' endonuclease assay and the ABI-PRISM 7900 for sequence detection (TaqMan). Initial quality control checks of the SNP assays were performed by the manufacturer (Applied Biosystems, Foster City, CA, USA); an additional 500 test reactions were run at the University of Southern California. Characteristics for the 10 TaqMan assays are available on a public website http://www.uscnorris.com/mecgenetics/CohortGCKView.aspx. Sequence validation for each SNP assay was performed on samples from the SNP500 project http://snp500cancer.nci.nih.gov[14] and 100% concordance was observed. To assess inter-laboratory variation, each genoty** center ran assays on a designated set of 94 samples from the Coriell Biorepository (Camden, NJ, USA) included in SNP500. The internal quality of genotype data at each genoty** center was assessed by ty** 5–10% blinded samples in duplicate or triplicate (depending on study).
Hormone Analysis
Circulating serum hormones were measured at the International Agency for Research on Cancer for EPIC and MEC samples and at the Harvard School of Public Health for NHS samples, for a total of 4713 subjects (1405 cases and 3308 controls, 1120 pre-menopausal and 3593 post-menopausal subjects). The different assays for hormone analyses were chosen on the basis of a previously published comparative validation study [15]. Estradiol (E2), estrone (E1) and androstenedione (Δ4) were measured by direct double-antibody radioimmunoassays from DSL (Diagnostic Systems Laboratories, Texas), while testosterone (T) was measured by direct radioimmunoassays from Immunotech (Marseille, France). Measurements were performed on never thawed serum sample aliquots. Mean intrabatch and interbatch coefficients of variation were 5.8 and 13.1%, respectively, for E2 (at a concentration of 250 pmol/l), 10.2 and 12.6% for E1 (at 75 pmol/l), and 4.8 and 18.9% for Δ4 (at 1.40 nmol/l), 10.8 and 15.3% for T (at 1.40 nmol/l).
Statistical Analysis
We used conditional multivariate logistic regression to estimate odds ratios (ORs) for invasive breast cancer in subjects with a linear (log-odds additive) scoring for 0, 1 or 2 copies of the minor allele of each SNP. We also used conditional logistic regression with additive scoring and the most common haplotype as the referent to estimate haplotype-specific ORs using an expectation-substitution approach to assign haplotypes based on the unphased genotype data and to account for uncertainty in assignment [16, 17]. Haplotype frequencies and expected subject-specific haplotype indicators were calculated separately for each cohort (and country within EPIC or ethnicity in the MEC). We combined rare haplotypes (those with estimated individual frequencies less than 3% in all cohorts) into a single category, which had a combined frequency of less than 1% of the controls for both genes and both linkage disequilibrium (LD) blocks of GNRHR. To test the global null hypothesis of no association between variation in GNRH1/GNRHR haplotypes and htSNPs and risk of invasive breast cancer (or subtypes defined by receptor status), we used a likelihood ratio test comparing a model with additive effects for each common haplotype (treating the most common haplotype as the referent) to the intercept-only model.
We performed subgroup analyses stratifying by cohort, ethnicity, country within EPIC, estrogen receptor/progesterone receptor status, metastatic vs. localized disease, and age at diagnosis (≤55 years vs. >55 years). We also investigated interactions between single SNPs or haplotypes and completion of a full term pregnancy (ever/never), age at first full term pregnancy (in three categories: nulliparous, ≤24, >24), body mass index (BMI in kg/m2 in three categories: <25, 25–29, ≥30), height (<160 cm, 160–165 cm, >165 cm), smoking status (never/former/current smoker), and use of menopausal hormone therapy (ever/never). Other common risk factors, including family history of breast cancer, personal history of benign breast disease, and age at menopause were unavailable for large numbers of women, and therefore were not included in the models.
Relationships of genetic variants with serum hormone levels were estimated by standard regression models, adjusted for BMI, age, assay batch, ethnicity, and country within EPIC. These analyses were performed both using all the study subjects for whom hormone levels have been measured, and only the controls, who represent the populations giving rise to the cases.
Results
The genomic regions surrounding GNRH1 and GNRHR are shown in Figure 1. GNRH1 consists of a single LD block, whereas GNRHR includes two LD blocks, one of them including exon 1 and the other exons 2 and 3. GNRH1 was tagged by 3 SNPs, which account for 94% of haplotype diversity. Block 1 of GNRHR was tagged by 3 SNPs and block 2 by 4 SNPs (98% and 95% of haplotype diversity, respectively). Frequency of common haplotypes ranged between 19 and 35% in controls for GNRH1 and 5% and 52% for GNRHR.
Haploview plot of the genomic region of GNRH1 (A) and GNRHR (B). From top to bottom: position of genes (boxes: exons, lines: introns), SNPs genotyped in the multiethnic panel, graphical representation of LD and block structure (darker color represents higher LD, numbers in the colored squares are percentage of LD, expressed as D', absence of number means D' = 100%).
A total of 5,603 invasive breast cancer cases and 7,480 controls were available for genoty** from each of the participating cohorts. Samples not yielding a genotype were removed from individual SNP analyses, and samples not yielding at least one genotype were removed from haplotype analyses. Both between-center genoty** concordance and within-center blinded quality control concordance were above 99%. Genotype success rate among cases and controls in all cohorts was greater than 95%. No polymorphisms deviated from Hardy-Weinberg Equilibrium among the controls.
Detailed results of associations between serum concentrations of steroid hormones and SNPs are presented in Additional file 1. SNP rs2630488 within GNRHR showed a nominally significant association (p = 0.04) with estradiol levels in post-menopausal women. Presence of the minor allele at this polymorphism was associated with an increase in estradiol level (a 4% increase among homozygotes for the minor allele, as compared to homozygotes for the common allele). However, this effect was entirely driven by the association observed in post-menopausal breast cancer cases within EPIC (p = 0.012 in this subgroup), and was not observed in the other subgroups. A few borderline associations were observed between hormone levels and SNPs in pre-menopausal women, however sample size of this group of subjects was considerably smaller, and all subjects derived from only one cohort (EPIC). No associations between polymorphisms and hormone levels remained significant after correction for multiple testing.
Results of association analyses between htSNPs of GNRH1 and breast cancer risk are presented in Table 1, and results of haplotype analysis in Table 2. Results of analyses for GNRHR are presented in Tables 3 and 4. No association was observed for any of the htSNPs of either gene. Haplotype analysis also showed no association, with global tests for comparison of haplotypes frequency in cases and controls resulting in non-significant results (Wald tests: d.f. = 4, p = 0.364 for GNRH1; d.f. = 5, p = 0.897 for GNRHR block 1 and d.f. = 6, p = 0.967 for GNRHR block 2). Analyses were unadjusted (conditional on matching criteria) or adjusted for known breast cancer risk factors, but results did not show any difference.
Analyses performed by stratifying cases by age at diagnosis (greater or lower than 55 years), localized or metastatic disease or estrogen/progesterone receptor status did not show significant differences. Stratification of subjects by cohort, country in EPIC or ethnicity in MEC showed only few results supported by p values ranging from 0.01 to 0.05, which were always based on a small number of subjects. For these, we performed heterogeneity tests, which in all cases were not statistically significant. For example, heterozygotes for SNP rs1812594 of GNRH1 had an odds ratio (OR) of 1.08 (95% confidence interval (CI) = 1.00–-1.16, p = 0.04). When we analyzed the results for each cohort, it resulted that the association was driven by EPIC data (OR = 1.16, 95% CI 1.00–1.34, p = 0.046), and within EPIC the only significant result came from the Spanish sub-cohort (OR = 1.59, 95% CI = 1.07–2.37, p = 0.021), which is based on 62 cases and 79 controls heterozygous for this SNP. However, heterogeneity tests for this genotype were not statistically significant for either the entire study (p = 0.226) or within EPIC (p = 0.147). Nor were homozygotes for the less common allele at this SNP significantly associated with increased risk in any subgroup.
No statistically significant interactions were observed between htSNPs or haplotypes and known breast cancer risk factors, including age at first full term pregnancy, number of pregnancies, never/ever menopausal hormone therapy, height, smoking status, or body mass index.
Discussion
This large, comprehensive study found no statistically significant associations between polymorphisms in the genes that code for GNRH1 or its receptor and either circulating ovarian sex hormones or breast cancer risk. An influence of SNPs in these two genes on breast cancer risk, mediated by altered levels of estrogens, was plausible, due to the known physiology of steroid hormone stimulation. If any common variants with functional relevance exist in the two candidate genes, our resequencing and haplotype tagging approaches should have detected its effect on hormone measurements and/or cancer risk. The null results are especially convincing because of the large sample size (more than 5,600 invasive breast cancer cases and 7,400 controls) and the extensive resequencing that preceded and informed the selection of htSNPs. The study has over 80% power to detect main effects of common polymorphisms (minor allele frequency of 5% or greater) with relative risks of 1.2 or greater, and to investigate interactions between genetic variants and known environmental or lifestyle exposures [18].
Based on these results, we conclude that common polymorphisms in GNRH1 and GNRHR do not substantially affect breast cancer risk in Caucasians. Among the many tests performed in subgroups, some associations were supported by p values ranging from 0.01 and 0.05, yet these associations were driven by subgroups containing small numbers of cases, and are therefore compatible with chance. None of these subgroup findings remain statistically significant when adjusted for multiple hypothesis testing. Likewise, the weakly significant association we observed between estradiol levels in post-menopausal women and SNP rs2630488 of GNRHR derived from a single subgroup in one cohort, and is also likely to reflect chance.
A limitation of this study was the relatively small number of subjects from racial or ethnic groups other than Caucasian. The MEC provided most of the cases and controls in this regard, but none of these groups exceeded 425 breast cancer cases. This limitation is particularly relevant to African Americans, for whom additional SNPs would be needed to provide comparable coverage of common variants. Coverage is satisfactory for the other ethnic groups [19]. This is consistent with genome-wide data [20], which also showed that tagging SNPs for Caucasians offer good coverage in other ethnic groups, except Africans.
Conclusion
In conclusion, we can exclude the possibility that common polymorphisms in GNRH1 and GNRHR confer large or even moderate breast cancer risks in Caucasians. We cannot exclude the possible existence of moderate risks due to polymorphisms of GNRH1 and GNRHR in non-Caucasian populations. Larger studies of non-Caucasians will be necessary to test this hypothesis.
References
Henderson BE, Feigelson HS: Hormonal carcinogenesis. Carcinogenesis. 2000, 21: 427-433. 10.1093/carcin/21.3.427.
Kaaks R, Rinaldi S, Key TJ, Berrino F, Peeters PH, Biessy C, Dossus L, Lukanova A, Bingham S, Khaw KT, Allen NE, Bueno-de-Mesquita HB, van Gils CH, Grobbee D, Boeing H, Lahmann PH, Nagel G, Chang-Claude J, Clavel-Chapelon F, Fournier A, Thiebaut A, Gonzalez CA, Quiros JR, Tormo MJ, Ardanaz E, Amiano P, Krogh V, Palli D, Panico S, Tumino R, et al: Postmenopausal serum androgens, oestrogens and breast cancer risk: the European prospective investigation into cancer and nutrition. Endocr Relat Cancer. 2005, 12: 1071-1082. 10.1677/erc.1.01038.
Eliassen AH, Missmer SA, Tworoger SS, Spiegelman D, Barbieri RL, Dowsett M, Hankinson SE: Endogenous steroid hormone concentrations and risk of breast cancer among premenopausal women. J Natl Cancer Inst. 2006, 98: 1406-1415.
Griffin JE, Ojeda SR, Eds: Textbook of endocrine physiology. 1996, New York: Oxford University Press
Barontini M, Garcia-Rudaz MC, Veldhuis JD: Mechanisms of hypothalamic-pituitary-gonadal disruption in polycystic ovarian syndrome. Arch Med Res. 2001, 32: 544-552. 10.1016/S0188-4409(01)00325-3.
Waldstreicher J, Seminara SB, Jameson JL, Geyer A, Nachtigall LB, Boepple PA, Holmes LB, Crowley WF: The genetic and clinical heterogeneity of gonadotropin-releasing hormone deficiency in the human. J Clin Endocrinol Metab. 1996, 81: 4388-4395. 10.1210/jc.81.12.4388.
Hunter DJ, Riboli E, Haiman CA, Albanes D, Altshuler D, Chanock SJ, Haynes RB, Henderson BE, Kaaks R, Stram DO, Thomas G, Thun MJ, Blanché H, Buring JE, Burtt NP, Calle EE, Cann H, Canzian F, Chen YC, Colditz GA, Cox DG, Dunning AM, Feigelson HS, Freedman ML, Gaziano JM, Giovannucci E, Hankinson SE, Hirschhorn JN, Hoover RN, Key T, et al: A candidate gene approach to searching for low-penetrance breast and prostate cancer genes. Nat Rev Cancer. 2005, 5: 977-985. 10.1038/nrc1754.
Calle EE, Rodriguez C, Jacobs EJ, Almon ML, Chao A, McCullough ML, Feigelson HS, Thun MJ: The American Cancer Society Nutrition Cohort: rationale, study design and baseline characteristics. Cancer. 2002, 94: 2490-2501. 10.1002/cncr.101970.
Riboli E, Hunt KJ, Slimani N, Ferrari P, Norat T, Fahey M, Charrondiere UR, Hemon B, Casagrande C, Vignat J, Overvad K, Tjønneland A, Clavel-Chapelon F, Thiebaut A, Wahrendorf J, Boeing H, Trichopoulos D, Trichopoulou A, Vineis P, Palli D, Bueno-De-Mesquita HB, Peeters PH, Lund E, Engeset D, Gonzalez CA, Barricarte A, Berglund G, Hallmans G, Day NE, Key TJ, et al: European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr. 2002, 5: 1113-1124. 10.1079/PHN2002394.
Colditz GA, Hankinson SE: The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer. 2005, 5: 388-396. 10.1038/nrc1608.
Rexrode K, Lee I, Cook N, Hennekens C, Buring J: Baseline characteristics of participants in the Women's Health Study. J Womens Health Gend Based Med. 2000, 9: 19-27. 10.1089/152460900318911.
Kolonel L, Henderson B, Hankin J, Nomura A, Wilkens L, Pike M, Stram D, Monroe K, Earle M, Nagamine F: A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000, 151: 346-357.
Stram DO, Haiman CA, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Pike MC: Choosing haplotype-tagging SNPS based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003, 55: 27-36. 10.1159/000071807.
Packer BR, Yeager M, Staats B, Welch R, Crenshaw A, Kiley M, Eckert A, Beerman M, Miller E, Bergen A, Rothman N, Strausberg R, Chanock SJ: SNP500Cancer: a public resource for sequence validation and assay development for genetic variation in candidate genes. Nucleic Acids Res. 2004, 32: D528-532. 10.1093/nar/gkh005.
Rinaldi S, Dechaud H, Biessy C, Morin-Raverot V, Toniolo P, Zeleniuch-Jacquotte A, Akhmedkhanov A, Shore RE, Secreto G, Ciampi A, Riboli E, Kaaks R: Reliability and validity of commercially available, direct radioimmunoassays for measurement of blood androgens and estrogens in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2001, 10: 757-765.
Zaykin D, Westfall P, Young S, Karnoub M, Wagner M, Ehm M: Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002, 53: 79-91. 10.1159/000057986.
Kraft P, Cox DG, Paynter R, Hunter D, De Vivo I: Accounting for haplotype uncertainty in association studies: A comparison of simple and flexible techniques. Genet Epidemiol. 2005, 28: 261-272. 10.1002/gepi.20061.
Garcia-Closas M, Rothman N, Lubin J: Misclassification in case-control studies of gene-environment interactions: assessment of bias and sample size. Cancer Epidemiol Biomarkers Prev. 1999, 8: 1043-1050.
Sedlmeyer IL, Pearce CL, Trueman JA, Butler JL, Bersaglieri T, Read AP, Clayton PE, Kolonel LN, Henderson BE, Hirschhorn JN, Palmert MR: Determination of sequence variation and haplotype structure for the gonadotropin-releasing hormone (GnRH) and GnRH receptor genes: investigation of role in pubertal timing. J Clin Endocrinol Metab. 2005, 90: 1091-1099. 10.1210/jc.2004-0649.
de Bakker PI, Burtt NP, Graham RR, Guiducci C, Yelensky R, Drake JA, Bersaglieri T, Penney KL, Butler J, Young S, Onofrio RC, Lyon HN, Stram DO, Haiman CA, Freedman ML, Zhu X, Cooper R, Groop L, Kolonel LN, Henderson BE, Daly MJ, Hirschhorn JN, Altshuler D: Transferability of tag SNPs in genetic association studies in multiple populations. Nat Genet. 2006, 38: 1298-1303. 10.1038/ng1899.
Pre-publication history
The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/9/257/prepub
Acknowledgements
We thank the participants in the component cohort studies. This work was funded by NCI grants U01 CA098216 (EPIC), U01CA098233 (Harvard), U01CA098758 (MEC) and U01 CA098710 (ACS). All co-authors of this paper are members of the NCI Breast and Prostate Cancer Cohort Consortium (BPC3).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FC, RK, DGC, KDH and BEH made up the writing committee for this work, and were responsible for data analyses, manuscript preparation and editing. CLP performed the htSNP selection and contributed substantially to manuscript editing. LD, CAH, DOS, SC provided expertise in genoty** and results analyses, as well as manuscript editing. All other authors contributed substantially to sample collection and manuscript editing. All authors read and approved the final version of the manuscript.
Electronic supplementary material
12885_2009_1591_MOESM1_ESM.xls
Additional file 1: Detailed results of associations between serum concentrations of steroid hormones and SNPs. (XLS 40 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Canzian, F., Kaaks, R., Cox, D.G. et al. Genetic polymorphisms of the GNRH1 and GNRHR genes and risk of breast cancer in the National Cancer Institute Breast and Prostate Cancer Cohort Consortium (BPC3). BMC Cancer 9, 257 (2009). https://doi.org/10.1186/1471-2407-9-257
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2407-9-257