Abstract
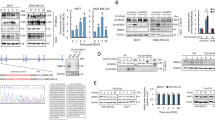
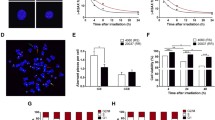
The p53 transcription factor is the most important sensor of ionizing radiation. Among the numerous effectors of p53 are the genes BBC3 and PMAIP1 encoding proapoptotic proteins PUMA and Noxa, respectively, as well as the CDKN1A/p21 cell cycle inhibitor. The effectiveness of radiation exposure—whether there is cell death or survival—is determined by the balance of mechanisms regulated by these proteins. In the present work, the predominant role was shown of BBC3/PUMA and CDKN1A/p21 versus PMAIP1/Noxa in p53-mediated responses to therapeutic doses of γ-radiation on isogenic human tumor cell lines (HCT116 colorectal carcinoma and HCT116p53KO subline with nonfunctioning p53). Bioinformatics analysis of genome-wide nucleotide sequences revealed significant differences in the putative motifs of p53 binding in the structure of genes BBC3 and PMAIP1. The results obtained are important for the development of targeted effects that would allow preserving the p53-dependent activation of proapoptotic genes while limiting the blocking of the cell cycle in irradiated tumor cells.




Similar content being viewed by others
REFERENCES
Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, vol. 72, p. 248.
Broude, E.V., Loncarek, J., Wada, I., Cole, K., Hanko, C., Roninson, I.B., and Swift, M., Mitotic catastrophe in cancer therapy, in Beyond Apoptosis: Cellular Outcomes of Cancer Therapy, New York: Informa Healthcare, 2008, p. 307.
Bunz, F., Dutriaux, A., Lengauer, C., Waldman, T., Zhou, S., Brown, J.P., and Vogelstein, B., Requirement for p53 and p21 to sustain G2 arrest after DNA damage, Science, 1998, vol. 282, p. 1497.
Chen, L., Willis, S.N., Wei, A., Smith, B.J., Fletcher, J.I., Hinds, M.G., and Huang, D.C., Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function, Mol. Cell, 2005, vol. 17, p. 393.
Davis, A.L., Qiao, S., Lesson, J.L., De La Vega, M.R., Park, S.L., Seanez, C.M., and Wondrak, G.T., The quinone methide aurin is a heat shock response inducer that causes proteotoxic stress and Noxa-dependent apoptosis in malignant melanoma cells, J. Biol. Chem., 2015, vol. 290, p. 1623.
Farré, D., Roset, R., Huerta, M., Adsuara, J.E., Roselló, L., Albà, M.M., and Messeguer, X., Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN, Nucleic Acids Res., 2003, vol. 31, p. 3651.
Fedele, M., Crescenzi, E., and Cerchia, L., The POZ/BTB and AT-hook containing zinc finger 1 (PATZ1) transcription regulator: physiological functions and disease involvement, Int. J. Mol. Sci., 2017, vol. 18, p. 2524.
Fernandez-Zapico, M.E., Lomberk, G.A., Tsuji, S., DeMars, C.J., Bardsley, M.R., Lin, Y.H., and Urrutia, R., A functional family-wide screening of SP/KLF proteins identifies a subset of suppressors of KRAS-mediated cell growth, Biochem. J., 2011, vol. 435, p. 529.
Gajjar, M., Candeias, M.M., Malbert-Colas, L., Ma-zars, A., Fujita, J., Olivares-Illana, V., and Fahraeus, R., The p53 mRNA-Mdm2 interaction controls Mdm2 nuclear trafficking and is required for p53 activation following DNA damage, Cancer Cell, 2012, vol. 21, p. 25.
Grant, C.E., Bailey, T.L., and Noble, W.S., FIMO: Scanning for occurrences of a given motif, Bioinformatics, 2011, vol. 27, p. 1017.
Hemann, M.T. and Lowe, S.W., The p53-BCL-2 connection, Cell Death Differ., 2006, vol. 13, p. 1256.
Huerta, S., Gao, X., Dineen, S., Kapur, P., Saha, D., and Meyer, J., Role of p53, Bax, p21, and DNA-PKcs in radiation sensitivity of HCT-116 cells and xenografts, Surgery, 2013, vol. 154, p. 143.
Kel, A.E., Gossling, E., Reuter, I., Cheremushkin, E., Kel-Margoulis, O.V., and Wingender, E., MATCHTM: a tool for searching transcription factor binding sites in DNA sequences, Nucleic Acids Res., 2003, vol. 31, p. 3576.
Kim, W., Lee, S., Seo, D., Kim, D., Kim, K., Kim, E., and Youn, B., Cellular stress responses in radiotherapy, Cells, 2019, vol. 8, p. 1105.
Kreis, N.N., Sanhaji, M., Rieger, M.A., Louwen, F., and Yuan, J., p21Waf1/Cip1 deficiency causes multiple mitotic defects in tumor cells, Oncogene, 2014, vol. 33, p. 5716.
Kuribayashi, K., Finnberg, N.K., Jeffers, J.R., Zambetti, G.P., and El-Deiry, W.S., The relative contribution of pro-apoptotic p53-target genes in the triggering of apoptosis following DNA damage in vitro and in vivo, Cell Cycle, 2011, vol. 10, p. 2380.
Lee, W.P., Lan, K.H., Li, C.P., Chao, Y., Lin, H.C., and Lee, S.D., Akt phosphorylates myc-associated zinc finger protein (MAZ), releases P-MAZ from the p53 promoter, and activates p53 transcription, Cancer Lett., 2016, vol. 375, p. 9.
Leibowitz, B.J., Qiu, W., Liu, H., Cheng, T., Zhang, L., and Yu, J., Uncoupling p53 functions in radiation-induced intestinal damage via PUMA and p21, Mol. Cancer Res., 2011, vol. 9, p. 616.
Maréchal, A. and Zou, L., DNA damage sensing by the ATM and ATR kinases, Cold Spring Harb. Perspect. Biol., 2013, vol. 5. https://doi.org/10.1101/cshperspect.a012716
Nakano, K. and Vousden, K.H., PUMA, a novel proapoptotic gene, is induced by p53, Mol. Cell, 2001, vol. 7, p. 683.
Nikiforov, M.A., Riblett, M., Tang, W.H., Gratchouck, V., Zhuang, D., Fernandez, Y., and Soengas, M.S., Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition, Proc. Natl. Acad. Sci. U. S. A., 2007, vol. 104, p. 19488.
Paiva, S.L. and Crews, C.M., Targeted protein degradation: Elements of PROTAC design, Curr. Opin. Chem. Biol., 2019, vol. 50, p. 111.
Ploner, C., Kofler, R., and Villunger, A., Noxa: At the tip of the balance between life and death, Oncogene, 2008, vol. 27, p. 84.
Rowland, B.D., Bernards, R., and Peeper, D.S., The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene, Nat. Cell. Biol., 2005, vol. 7, p. 1074.
Speidel, D., The role of DNA damage responses in p53 biology, Arch. Toxicol., 2015, vol. 89, p. 501.
Valentino, T., Palmieri, D., Vitiello, M., Pierantoni, G.M., Fusco, A., and Fedele, M., PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context, Cell Death Dis., 2013, vol. 4. https://doi.org/10.1038/cddis.2013.500
Vavrova, J. and Rezacova, M., Importance of proapoptotic protein PUMA in cell radioresistance, Folia Biol., 2014, vol. 60, p. 53.
Wang, Q., Mora-Jensen, H., Weniger, M.A., Perez-Galan, P., Wolford, C., Hai, T., and Ye, Y., ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells, Proc. Natl. Acad. Sci. U. S. A., 2009, vol. 106, p. 2200.
Zhu, N., Gu, L., Findley, H.W., Chen, C., Dong, J.T., Yang, L., and Zhou, M., KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia, J. Biol. Chem., 2006, vol. 281, p. 14711.
Funding
This study was financially supported by the Russian Foundation for Basic Research, project no. 34-90046.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare they have no conflict of interest. The work does not contain any research using animals or human beings as objects of research.
Additional information
Abbreviations: BBC3—Bcl-2 binding component 3, CDKN1A—cyclin-dependent kinase 1A, PMAIP1—phorbol-12-myristate-13-acetate-inducible protein 1, PUMA—p53-regulated (up-regulated) modulator of apoptosis.
Rights and permissions
About this article
Cite this article
Kuchur, O.A., Kuchur, P.D., Kuzmina, D.O. et al. Differential Regulation of BBC3/PUMA and PMAIP1/Noxa in Ionizing Radiation: the Role of p53. Cell Tiss. Biol. 15, 544–553 (2021). https://doi.org/10.1134/S1990519X21060043
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990519X21060043