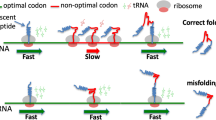
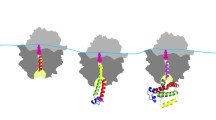
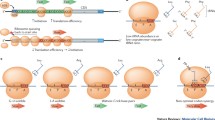
Abstract—In the cell, protein folding begins during protein synthesis/translation and thus is a co-translational process. Co-translational protein folding is tightly linked to translation elongation, which is not a uniform process. While there are many reasons for translation non-uniformity, it is generally believed that non-uniform synonymous codon usage is one of the key factors modulating translation elongation rates. Frequent/optimal codons as a rule are translated more rapidly than infrequently used ones and vice versa. Over 30 years ago, it was hypothesized that changes in synonymous codon usage affecting translation elongation rates could im**e on co-translation protein folding and that many synonymous codons are strategically placed within mRNA to ensure a particular translation kinetics facilitating productive step-by-step co-translational folding of proteins. It was suggested that this particular translation kinetics (and, specifically, translation pause sites) may define the window of opportunity for the protein parts to fold locally, particularly at the critical points where folding is far from equilibrium. It was thus hypothesized that synonymous codons may provide a secondary code for protein folding in the cell. Although, mostly accepted now, this hypothesis appeared to be difficult to prove and many convincing results were obtained only relatively recently. Here, I review the progress in the field and explain, why this simple idea appeared to be so challenging to prove.



Similar content being viewed by others
REFERENCES
Hartl F.U. 2017. Protein misfolding diseases. Annu. Rev. Biochem.86, 21‒26.
Chiti F., Dobson C.M. 2017. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem.86, 27‒68.
Soto C., Pritzkow S. 2018. Protein misfolding, aggregation, and conformational strains in neurodegenerative diseases. Nat. Neurosci.21, 1332‒1340.
Fersht A.R. 2008. From the first protein structures to our current knowledge of protein folding: Delights and skepticisms. Nat. Rev. Mol. Cell Biol.9, 650‒654.
Ferina J., Daggett V. 2019. Visualizing protein folding and unfolding. J. Mol. Biol.431, 1540–1564.
Anfinsen C.B. 1973. Principles that govern the folding of protein chains. Science.181, 223‒230.
Finkelstein A.V. 2018. 50+ years of protein folding. Biochemistry (Moscow). 83 (Suppl. 1), S3‒S18.
Bartlett A.I., Radford S.E. 2009. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat. Struct. Mol. Biol.16, 582‒588.
Abaskharon R.M., Gai F. 2016. Meandering down the energy landscape of protein folding: Are we there yet? Biophys. J.110, 1924‒1932.
Jaenicke R. 1991. Protein folding: local structures, domains, subunits, and assemblies. Biochemistry.30, 3147‒3161.
Hartl F.U., Hayer-Hartl M. 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol.16, 574‒581.
Hingorani K.S. Gierasch L.M. 2014. Comparing protein folding in vitro and in vivo: Foldability meets the fitness challenge. Curr. Opin. Struct. Biol.24, 81‒90.
Balchin D., Hayer-Hartl M., Hartl F.U. 2016. In vivo aspects of protein folding and quality control. Science. 353, aac4354.
Gruebele M., Dave K., Sukenik S. 2016. Globular protein folding in vitro and in vivo. Annu. Rev. Biophys.45, 233‒251.
Dahiya V., Buchner J. 2019. Functional principles and regulation of molecular chaperones. Adv. Protein Chem. Struct. Biol.114, 1‒60.
Jayaraj G.G., Hipp M.S., Hartl F.U. 2019. Functional modules of the proteostasis network. Cold Spring Harb. Perspect. Biol. Mar. 4, pii: a033951. https://doi.org/10.1101/cshperspect.a033951
Komar A.A. 2009. A pause for thought along the co-translational folding pathway. Trends Biochem. Sci.34, 16‒24.
Kramer G., Boehringer D., Ban N., Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol.16, 589‒597.
Cabrita L.D., Dobson C.M., Christodoulou J. 2010. Protein folding on the ribosome. Curr. Opin. Struct. Biol.20, 33‒45.
Pechmann S., Willmund F., Frydman J. 2013. The ribosome as a hub for protein quality control. Mol. Cell.49, 411‒421.
Gloge F. Becker A.H. Kramer G., Bukau B. 2014. Co-translational mechanisms of protein maturation. Curr. Opin. Struct. Biol.24, 24‒33.
Chaney J.L., Clark P.L. 2015. Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys.44, 143‒166.
Thommen M., Holtkamp W., Rodnina M.V. 2017. Co-translational protein folding: Progress and methods. Curr. Opin. Struct. Biol.42, 83‒89.
Komar A.A. 2018. Unraveling co-translational protein folding: Concepts and methods. Methods.137, 71‒81.
Williams N.K, Dichtl B. 2018. Co-translational control of protein complex formation: A fundamental pathway of cellular organization? Biochem. Soc. Trans.46, 197‒206.
Cowie D.B., Spiegelman S., Roberts R.B., Duerksen J.D. 1961. Ribosome-bound beta-galactosidase. Proc. Natl. Acad. Sci. U. S. A.47, 114‒122.
Zipser D., Perrin D. 1963. Complementation on ribosomes. Cold Spring Harbor. Symp. Quant. Biol.28, 533‒537.
Kiho Y., Rich A. 1964. Induced enzyme formed on bacterial polyribosomes. Proc. Natl. Acad. Sci. U. S. A.51, 111‒118.
Hamlin J., Zabin I. 1972. β-Galactosidase: Immunological activity of ribosome-bound, growing polypeptide chains. Proc. Natl. Acad. Sci. U. S. A.69, 412‒416.
Bergman L.W., Kuehl W.M. 1979. Formation of intermolecular disulfide bonds on nascent immunoglobulin polypeptides. J. Biol. Chem.254, 5690‒5694.
Bergman L.W., Kuehl W.M. 1979. Formation of an intrachain disulfide bond on nascent immunoglobulin light chains. J. Biol. Chem.254, 8869‒8876.
Bergman L.W., Kuehl W.M. 1979. Co-translational modification of nascent immunoglobulin heavy and light chains. J. Supramol. Struct.11, 9‒24.
Gilbert R.J., Fucini P., Connell S., Fuller S.D., Nierhaus K.H., Robinson C.V., Dobson C.M., Stuart D.I. 2004. Three-dimensional structures of translating ribosomes by Cryo-EM. Mol. Cell.14, 57‒66.
Kosolapov A., Deutsch C. 2009. Tertiary interactions within the ribosomal exit tunnel. Nat. Struct. Mol. Biol.16, 405‒411.
Tu L., Khanna P., Deutsch C. 2014. Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J. Mol. Biol.426, 185‒198.
Holtkamp W., Kokic G., Jäger M., Mittelstaet J., Komar A.A., Rodnina M.V. 2015. Cotranslational protein folding on the ribosome monitored in real time. Science. 350, 1104‒1107.
Komar A.A. 2018. The Yin and Yang of codon usage. Hum. Mol. Genet.25 (R2), R77–R85.
Schuller A.P., Green R. 2018. Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol.19, 526‒541.
Purvis I.J., Bettany A.J., Santiago T.C., Coggins J.R., Duncan K., Eason R., Brown A.J. 1987. The efficiency of folding of some proteins is increased by controlled rates of translation in vivo. A hypothesis. J. Mol. Biol.193, 413‒417.
Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. 1988. Role of the rare codon clusters in defining the boundaries of polypeptide chain regions with identical secondary structures in the process of co-translational folding of proteins. Dokl. Akad. Nauk SSSR.303, 995‒999.
Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. 1989. Frequency of using codons in mRNA and coding of the domain structure of proteins. Dokl. Akad. Nauk SSSR.305, 1006‒1012.
Krasheninnikov I.A., Komar A.A., Adzhubeĭ I.A. 1989. Role of the code redundancy determining cotranslational protein folding. Biokhimiya.5, 187‒200.
Crick F.H., Barnett L., Brenner S., Watts-Tobin R.J. 1961. General nature of the genetic code for proteins. Nature. 192, 1227‒1232.
Hershberg R., Petrov D.A. 2008. Selection on codon bias. Annu. Rev. Genet.42, 287‒299.
Sharp P.M., Emery L.R. Zeng K. 2010. Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. Lond. B.365, 1203‒1212.
Behura S.K.m Severson D.W. 2013. Codon usage bias: Causative factors, quantification methods and genome-wide patterns, with emphasis on insect genomes. Biol. Rev. Camb. Philos. Soc.88, 49‒61.
Ikemura T. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol.2, 13‒34.
Sharp P.M., Cowe E., Higgins D.G., Shields D.C., Wolfe K.H., Wright F. 1988. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens: A review of the considerable within-species diversity. Nucleic Acids Res.16, 8207‒8211.
Andersson S.G., Kurland C.G. 1990. Codon preferences in free-living microorganisms. Microbiol. Rev.,54, 198‒210.
Nakamura Y., Gojobori T., Ikemura T. 2000. Codon usage tabulated from the international DNA sequence databases: Status for the year 2000. Nucleic Acids Res.28, 292.
Ikemura T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol.146, 1‒21.
Ikemura T. 1982. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J. Mol. Biol.158, 573‒597.
Quax T.E., Claassens N.J., Söll D., van der Oost J. 2015. Codon bias as a means to fine-tune gene expression. Mol. Cell.59, 149‒161.
Komar A.A. 2016. The art of gene redesign and recombinant protein production: approaches and perspectives. In: Protein Therapeutics. Eds Zuben E. Sauna, Chava Kimchi-Sarfaty. Springer, pp 161‒177. doi.org/https://doi.org/10.1007/978-3-319-41818-6
Hanson G., Coller J. 2018. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol.19, 20‒30.
Sharp P.M., Tuohy T.M., Mosurski K.R. 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res.14, 5125‒5143.
Shields D.C., Sharp P.M. 1987. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res.15, 8023‒8040.
Sharp P.M., Devine K.M. 1989. Codon usage and gene expression level in Dictyostelium discoideum: Highly expressed genes do “prefer” optimal codons. Nucleic Acids Res.17, 5029‒5039.
Karlin S., Mrázek J., Campbell A.M. 1998. Codon usages in different gene classes of the Escherichia coli genome. Mol. Microbiol.29, 1341‒1355.
Brinkmann U., Mattes R.E., Buckel P. 1989. High-level expression of recombinant genes in Escherichia coli is dependent on the availability of the dnaY gene product. Gene.85, 109‒114.
Chen K.S., Peters T.C., Walker J.R. 1990. A minor arginine tRNA mutant limits translation preferentially of a protein dependent on the cognate codon. J. Bacteriol.172, 2504‒2510.
Chen G.T., Inouye M. 1994. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli.Genes Dev.8, 2641‒2652.
Zahn K., Landy A. 1996. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol. Microbiol.21, 69‒76.
Del Tito B.J., Jr., Ward J.M., Hodgson J., Gershater C.J., Edwards H., Wysocki L.A., Watson F.A., Sathe G., Kane J.F. 1995. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli.J. Bacteriol.177, 7086‒7091.
Protzel A., Morris A.J. 1974. Gel chromatographic analysis of nascent globin chains. Evidence of nonuniform size distribution. J. Biol. Chem.249, 4594‒4600.
Chaney W.G., Morris A.J. 1978. Nonuniform size distribution of nascent peptides: The role of messenger RNA. Arch. Biochem. Biophys.191, 734‒741.
Krasheninnikov I.A. Komar A.A., Adzhubeĭ I.A. 1991. Nonuniform size distribution of nascent globin peptides, evidence for pause localization sites, and a cotranslational protein-folding model. J. Protein Chem.10, 445–454.
Komar A.A., Jaenicke R. 1995. Kinetics of translation of gamma B crystallin and its circularly permutated variant in an in vitro cell-free system: Possible relations to codon distribution and protein folding. FEBS Lett.376, 195‒198.
Wolin S.L., Walter P. 1988. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J.7, 3559‒3569.
Hollingsworth M.J., Kim J.K., Stollar N.E. 1998. Heelprinting analysis of in vivo ribosome pause sites. Methods Mol. Biol.77, 153‒165.
Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science.324, 218‒223.
McGlincy N.J., Ingolia N.T. 2017. Transcriptome-wide measurement of translation by ribosome profiling. Methods.126, 112‒129.
Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb. Perspect. Biol.11 (5), pii: a032698. https://doi.org/10.1101/cshperspect.a032698
Hussmann J.A., Patchett S., Johnson A., Sawyer S., Press W.H. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet.11, e1005732.
Mohammad F., Green R., Buskirk A.R. 2019. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. eLife.8, e42591.
Kimura M. 1984. The Neutral Theory of Molecular Evolution Cambridge. Cambridge, UK: Cambridge Univ. Press.
Zhao Z., Fu Y.X., Hewett-Emmett D., Boerwinkle E. 2003. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene.312, 207‒213.
Chamary J.V., Parmley J.L., Hurst L.D. 2006. Hearing silence: Non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet.7, 98‒108.
Yarus M., Folley L.S. 1985. Sense codons are found in specific contexts. J. Mol. Biol.182, 529‒540.
Buckingham R.H. 1990. Codon context. Experientia. 46, 1126‒1133.
Buckingham, R.H. 1994. Codon context and protein synthesis: enhancements of the genetic code. Biochimie. 76, 351‒354.
Gutman G.A., Hatfield G.W. 1989. Nonrandom utilization of codon pairs in Escherichia coli.Proc. Natl. Acad. Sci. U. S. A.86, 3699‒3703.
Tats A., Tenson T., Remm M. 2008. Preferred and avoided codon pairs in three domains of life. BMC Genomics. 9, 463.
Diambra L.A. 2017. Differential bicodon usage in lowly and highly abundant proteins. PeerJ. 5, e3081.
Brule C.E., Grayhack E.J. 2017). Synonymous codons: Choose wisely for expression. Trends Genet.33, 283‒297.
Alexaki A., Kames J.M., Holcomb D.D., Athey J., Santana-Quintero L.V., Lam P.V., Hamasaki-Katagiri N., Osipova E., Simonyan V., Bar H., Komar A.A., Kimchi-Sarfaty C. 2019. Codon and Codon-Pair Usage Tables (CoCoPUTs): Facilitating genetic variation analyses and recombinant gene design. J. Mol. Biol.431 (13), 2434‒2441. https://doi.org/10.1016/j.jmb.2019.04.021
Képès F. 1996. The “+70 pause”: Hypothesis of a translational control of membrane protein assembly. J. Mol. Biol.262, 77‒86.
Clarke T.F., 4th, Clark P.L. 2008. Rare codons cluster. PLoS One.3, e3412.
Kudla G., Murray A.W., Tollervey D., Plotkin J.B. 2009. Coding-sequence determinants of gene expression in Escherichia coli.Science.324, 255‒258.
Clarke T.F., 4th, Clark P.L. 2010. Increased incidence of rare codon clusters at 5' and 3' gene termini: Implications for function. BMC Genomics.11, 118.
Goodman D.B., Church G.M., Kosuri S. 2013. Causes and effects of N-terminal codon bias in bacterial genes. Science.342, 475‒479.
Bentele K., Saffert P., Rauscher R., Ignatova Z., Bluthgen N. 2013. Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol.9, 675.
Pechmann S., Chartron J.W., Frydman J. 2014. Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo.Nat. Struct. Mol. Biol.21, 1100‒1105.
Thanaraj T.A., Argos P. 1996. Ribosome-mediated translational pause and protein domain organization. Protein Sci.5, 1594‒1612.
Thanaraj T.A., Argos P. 1996. Protein secondary structural types are differentially coded on messenger RNA. Protein Sci.5, 1973‒1983.
Adzhubei A.A., Adzhubei I.A., Krasheninnikov I.A., Neidle S. 1996. Non-random usage of 'degenerate' codons is related to protein three-dimensional structure. FEBS Lett.399, 78‒82.
Oresic M., Shalloway D. 1998. Specific correlations between relative synonymous codon usage and protein secondary structure. J. Mol. Biol.281, 31‒48.
Chartier M., Gaudreault F., Najmanovich R. 2012. Large-scale analysis of conserved rare codon clusters suggests an involvement in co-translational molecular recognition events. Bioinformatics. 28, 1438‒1445.
Widmann M., Clairo M., Dippon J., Pleiss J. 2008. Analysis of the distribution of f unctionally relevant rare codons. BMC Genomics.9, 207.
McKownvR.L., Raab R.W., Kachelries P., Caldwell S., Laurie G.W. 2013. Conserved regional 3' grou** of rare codons in the coding sequence of ocular prosecretory mitogen lacritin. Invest. Ophthalmol. Vis. Sci.54, 1979‒1987.
Gustafsson C., Govindarajan S., Minshull J. 2004. Codon bias and heterologous protein expression. Trends Biotechnol.22, 346–353.
Wu G., Zheng Y., Qureshi I., Zin H.T., Beck T., Bulka B., Freeland S.J. 2007. SGDB: A database of synthetic genes re-designed for optimizing protein over-expression. Nucleic Acids Res.35, D76–D79.
Quax T.E., Claassens N.J., Soll D., van der Oost J. 2015. Codon bias as a means to fine-tune gene expression. Mol. Cell. 59, 149–161.
Presnyak V., Alhusaini N., Chen Y.H., Martin S., Morris N., Kline N., Olson S., Weinberg, D., Baker K.E., Graveley B.R., Coller J. 2015. Codon optimality is a major determinant of mRNA stability. Cell.160, 1111‒1124.
Boël G., Letso R., Neely H., Price W.N., Wong K.H., Su M., Luff J.D., Valecha M., Everett J.K., Acton T.B., **ao R., Montelione G.T., Aalberts D.P., Hunt J.F. 2016. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature.529, 358‒363.
Mishima Y., Tomari Y. 2016. Codon usage and 3' UTR length determine maternal mRNA stability in zebrafish. Mol. Cell.61, 874‒885.
Sharp P.M., Li W.H. 1987. The Codon Adaptation Index: A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281‒1295.
Komar A.A., Lesnik T., Reiss C. 1999. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett.462, 387‒391.
Uemura S., Aitken C.E., Korlach J., Flusberg B.A., Turner S.W., Puglisi J.D. 2010. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature.464, 1012‒1017.
Prabhakar A., Puglisi E.V., Puglisi J.D. 2019. Single-molecule fluorescence applied to translation. Cold Spring Harb. Perspect. Biol.11, pii: a032714.
Buhr F., Jha S., Thommen M., Mittelstaet J., Kutz F., Schwalbe H., Rodnina M.V., Komar A.A. 2016. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell.61, 341‒351.
Komar A.A., Kommer A., Krasheninnikov I.A., Spirin A.S. 1993. Cotranslational heme binding to nascent globin chains. FEBS Lett.326, 261‒263.
Komar A.A., Kommer A., Krasheninnikov I.A., Spirin A.S. 1997. Cotranslational folding of globin. J. Biol. Chem.272, 10646‒10651.
Kolb V.A., Makeyev E.V., Spirin A.S. (1994). Folding of firefly luciferase during translation in a cell-free system. EMBO J.13, 3631‒3637.
Makeyev E.V., Kolb V.A., Spirin A.S. 1996. Enzymatic activity of the ribosome-bound nascent polypeptide. FEBS Lett.378, 166‒170.
Kimchi-Sarfaty C., Oh J.M., Kim I.W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science.315, 525‒528.
Zhang G., Hubalewska M., Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat. Struct. Mol. Biol.16, 274‒280.
Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J.M., Sachs M.S., Liu Y. 2013. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature.495, 111‒115.
Sander I.M., Chaney J.L., Clark P.L. 2014. Expanding Anfinsen’s principle: Contributions of synonymous codon selection to rational protein design, J. Am. Chem. Soc.136, 858‒861.
Hu S., Wang M., Cai G., He M. 2013. Genetic code-guided protein synthesis and folding in Escherichia coli.J. Biol. Chem.288, 30855‒30861.
Kim S.J., Yoon J.S., Shishido H., Yang Z., Rooney L.A., Barral J.M., Skach W.R. 2015. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science.348, 444‒448.
Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y. 2015. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell.59, 744‒754.
Komar A.A. 2007. SNPs, silent but not invisible. Science.315, 466‒467.
Komar A.A. 2007. Silent SNPs: Impact on gene function and phenotype. Pharmacogenomics.8, 1075–1080.
Sauna. Z.E., Kimchi-Sarfaty C. 2011. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet.12, 683‒691.
Hunt R.C., Simhadri V.L., Iandoli M., Sauna Z.E., Kimchi-Sarfaty C. 2014. Exposing synonymous mutations. Trends Genet.30, 308‒321.
Simhadri V.L., Hamasaki-Katagiri N., Lin B.C., Hunt R., Jha S., Tseng S.C., Wu A., Bentley A.A., Zichel R., Lu Q., Zhu L., Freedberg D.I., Monroe D.M., Sauna Z.E., Peters R., Komar A.A., Kimchi-Sarfaty C. 2017. Single synonymous mutation in factor IX alters protein properties and underlies haemophilia B. J. Med. Genet.54, 338‒345.
Knobe K.E., Sjorin E., Ljung R.C. 2008. Why does the mutation G17736A/Val107Val (silent) in the F9 gene cause mild haemophilia B in five Swedish families? Haemophilia. 14, 723–728.
Shyu Y.J., Liu H., Deng X., Hu C.D. 2006. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. Biotechniques.40, 61‒66.
Shyu Y.J., Hu C.D. 2008. Fluorescence complementation: An emerging tool for biological research. Trends Biotechnol.26, 622‒630.
Chen B., Kaledhonkar S., Sun M., Shen B., Lu Z., Barnard D., Lu T.M., Gonzalez R.L., Jr., Frank J. 2015. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure. 23, 1097‒1105.
ACKNOWLEDGMENTS
This work would not be possible without the original contribution of Ivan A. Adzhubei and Igor A. Krasheninnikov, further support from Slava Kolb, Aigar Kommer, Lev P. Ovchinnikov and Alexander S. Spirin, followed by collaborations with Rainer Jaenicke, Claude Reiss and more recently with Chava Kimchi-Sarfaty, Harald Schwalbe and Marina V. Rodnina.
I am indebted to all my colleagues and collaborators for their extremely generous and inspiring discussions and invaluable contributions.
I also apologize to those whose work or original publications could not be cited in this short review article.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
FUNDING
In recent years, this work in my laboratory was supported by grants from the Human Frontier Science Program Organization [HFSP grant #RGP0024/2010], the American Heart Association [AHA grant 13GRNT17070025], the National Institutes of Health [NIH grant HL121779], the Center for Gene Regulation in Health and Disease (GRHD) at CSU, and the biotechnology company, DAPCEL, Inc, that for more that 10 years has been successfully utilizing ideas developed during this study for synonymous gene optimization aimed at production (in any desired host organism) of correctly folded, soluble proteins.
COMPLIANCE WITH ETHICAL STANDARDS
The author declares that he has no conflict of interest. This article does not contain any studies involving animals or human participants performed by the author.
ADDITIONAL INFORMATION
The text was submitted by the author(s) in English.
Rights and permissions
About this article
Cite this article
Komar, A.A. Synonymous Codon Usage—a Guide for Co-Translational Protein Folding in the Cell. Mol Biol 53, 777–790 (2019). https://doi.org/10.1134/S0026893319060098
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893319060098