Abstract
Nasopalatine duct cysts are difficult to detect on panoramic radiographs due to obstructive shadows and are often overlooked. Therefore, sensitive detection using panoramic radiography is clinically important. This study aimed to create a trained model to detect nasopalatine duct cysts from panoramic radiographs in a graphical user interface-based environment. This study was conducted on panoramic radiographs and CT images of 115 patients with nasopalatine duct cysts. As controls, 230 age- and sex-matched patients without cysts were selected from the same database. The 345 pre-processed panoramic radiographs were divided into 216 training data sets, 54 validation data sets, and 75 test data sets. Deep learning was performed for 400 epochs using pretrained-LeNet and pretrained-VGG16 as the convolutional neural networks to classify the cysts. The deep learning system's accuracy, sensitivity, and specificity using LeNet and VGG16 were calculated. LeNet and VGG16 showed an accuracy rate of 85.3% and 88.0%, respectively. A simple deep learning method using a graphical user interface-based Windows machine was able to create a trained model to detect nasopalatine duct cysts from panoramic radiographs, and may be used to prevent such cysts being overlooked during imaging.
Similar content being viewed by others
Introduction
Since artificial intelligence surpassed human image classification accuracy at the ImageNet Large Scale Visual Recognition Challenge in 2015, artificial intelligence (AI) in the field of image diagnosis has begun to reach a practical level1. In particular, neural networks created with convolutional and pooling layers based on human vision, called convolutional neural networks, boast extremely high accuracy in image recognition2,3. Consequently, deep learning using convolutional neural networks has been actively performed in the medical radiology field4,5,6,7,8,9. Furthermore, for the maxillofacial region, active artificial intelligence research has begun to detect diseases and classify images using the panoramic radiographs that are routinely performed10,11,12,13,14,15,16.
A nasopalatine duct cyst (NPDC) is a develo** cyst that arises in the nasopalatine duct and is the most common non-odontogenic cyst17,18. NPDCs are thought to arise from the residual epithelium within the nasopalatine duct and are observed in all age groups17,18. Although NPDCs cause neurological symptoms, pain, and swelling, an early NPDC is often asymptomatic and early detection depends on radiographic imaging during routine clinical practice17,18,19. However, NPDCs are often difficult to detect on panoramic radiographs due to obstructive shadows, and are often overlooked. Furthermore, although rare, squamous cell carcinoma may arise in the epithelium of NPDCs20. Therefore, sensitive detection of NPDCs using panoramic radiography is clinically important.
Currently, there are few studies using panoramic radiograph to detect NPDC, and for the development of AI, it is essential to acquire artificial intelligence parameters at multiple facilities21. In addition, past studies have not used computed tomography (CT) to annotate training data, thus it is possible that parameters of NPDC which are difficult to detect with panoramic radiography, have not been obtained21. Moreover, conducting deep learning in a character user interface environment using Linux, which is currently widely used, is a barrier among researchers. There is a need for a simple method that can be used for widespread artificial intelligence development in the future.
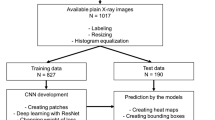
Therefore, this research aimed to create a trained model to detect NPDCs from panoramic radiographs with a graphical user interface-based environment using a Windows machine.
Materials and methods
This study was approved by the Ethics Committee of the University School of Dentistry (No. EC15-12-009-1). The requirement to obtain written informed consent was waived by the Ethics Committee for this retrospective study. All procedures followed the guidelines of the Declaration of Helsinki: ethical principles for medical research involving human subjects.
Subjects
This study was conducted on the panoramic radiographs and CT images of 115 patients with NPDCs (71 males, 44 females; mean age 46.7 ± 16.4, range 17–84) from April 2006 to April 2022. NPDCs were diagnosed by an oral and maxillofacial radiologist on CT or identified by histopathological examination of tissues excised during surgery. If the nasopalatine duct was enlarged by 6 mm or greater, and had expanded more than the incisive foramen and nasopalatine foramen on CT, an NPDC was diagnosed (Fig. 1)22. As controls, 230 age- and sex-matched patients without NPDCs (142 males, 88 females; mean age 46.7 ± 16.4, range 17–84) were selected from the same database. The case and control groups were patients who underwent panoramic radiography and CT for suspected jaw bone lesions. Patients with lesions in the maxillary anterior region were excluded from the control group.
Identification of nasopalatine duct cyst (NPDC) using computed tomography (CT). (a) Axial CT shows normal NPD (arrow). The maximum diameter of NPD is 5.0 mm. (b) Axial CT shows NPDC (arrow). The maximum diameter of NPD is 11.8 mm. If the maximum diameter of NPD was 6.0 mm or more on Axial CT, it was identified as NPDC.
Data preprocessing
CT imaging was performed with a 64-multidetector row CT system (Aquilion 64; Toshiba Medical Systems, Tokyo, Japan). All patients were scanned using the routine clinical protocol for craniomaxillofacial examination at our hospital, which was as follows: tube voltage, 120 kV; tube current, 100 mA; field of view, 240 × 240 mm; helical pitch, 41. The imaging included axial (0.50 mm), multiplanar (3.00 mm), and three-dimensional images. The CT images were interpreted using a medical liquid crystal display monitor (RadiForce G31; Eizo Nanao, Ishikawa, Japan).
All panoramic radiographs were taken using a panoramic radiography (Veraviewepocs: J Morita, Kyoto, Japan) examination at 1–10 mA with a peak voltage between 60 and 80 kV, depending on the patients’ jaw size. All panoramic radiographs were extracted as Joint Photographic Experts Group files.
To increase the NPDC detection accuracy of the convolutional neural network models, the maxillary anterior region was set as the ROI. Therefore, one radiologist manually segmented the panoramic radiograph images horizontally from the right to the left end of both the maxillary canines or lateral incisors, and vertically from the nasal floor to the mandibular anterior incisal edge level. If there were no teeth in the maxillary anterior region, the corresponding region was set as the ROI. The images were then saved as Joint Photographic Experts Group files (Fig. 2).
Dataset
The data set was prepared in comma separated values file format, with an 8-bit grayscale image and a matrix size of 256 × 256 pixels. The 345 preprocessed images were divided into 216 training data sets (71 in the NPDC group and 145 in the non-NPDC group), 54 validation data sets (19 in the NPDC group and 35 in the non-NPDC group), and 75 test data sets (25 in the NPDC group and 50 in the non-NPDC group).
Deep learning
To construct the NPDC predictive machine learning model, a Windows PC with an NVIDIA GeForce RTX 3090, and Neural Network Console version 2.10 (Sony Corp., Tokyo, Japan) were used as a deep learning-integrated development environment.
Deep learning was performed for 400 epochs using pretrained-LeNet and pretrained-VGG16 as convolutional neural networks to classify NPDCs. The optimization algorithm employed was the Adam optimizer, at a learning rate of 0.001, a weight decay of 0, a batch size of 32, and with batch normalization. Furthermore, the Train data was augmented to 86,400 data, which is 400 times (scale: 0.8–1.2, angle: 0.2, aspect ratio: 1.2, brightness: 0.02, contrast: 1.2, Flip LR: presence). These parameters were optimized and determined through preliminary experiments.
Diagnostic performance
The diagnostic performance was calculated for the testing set. The accuracy, sensitivity, and specificity, of the deep learning system using LeNet and VGG16, were calculated. Furthermore, a radiologist (with 6 years of experience) and a radiology specialist (with 11 years of experience) performed the test. Then, the artificial intelligence focal points of LeNet and VGG16 were visually evaluated using Gradient-weighted Class Activation Map** and Locally Interpretable Model-agnostic Explanations.
Results
Table 1 lists the diagnostic performance of the deep learning system using LeNet and VGG16. LeNet showed an accuracy rate of 85.3% (true positive 18, true negative 46, false positive 7, false negative 4) and LeNet showed an accuracy rate of 88.0% (true positive 19, true negative 47, false positive 6, false negative 3) in the test data. Moreover, in the learning curve, neither LeNet nor VGG16 showed a tendency of obvious overfitting (Fig. 3). However, LeNet showed a learning curve in which the validation error did not converge to the optimal solution.
Figure 4 presents examples of cropped panoramic radiographs and their corresponding Gradient-weighted Class Activation Map** and Locally Interpretable Model-agnostic Explanations images generated from deep learning-based NPDC detection models. Gradient-weighted Class Activation Map** showed that LeNet classified by focusing on the region corresponding to the nasopalatine duct. Grad-CAM showed that VGG16 focused on a relatively wide range of regions compared to LeNet, and focused on the edge of the NPDC. Locally Interpretable Model-agnostic Explanations indicated that LeNet focused on a specific part of the image each time. In contrast, VGG16 changed the focal point of the image each time. From Gradient-weighted Class Activation Map** and Locally Interpretable Model-agnostic Explanations, the deep learning-based NPDC detection models were not affected by an oral environment featuring tooth loss or dental materials.
Examples of cropped panoramic radiographs and their corresponding Gradient-weighted Class Activation Map** and Locally Interpretable Model-agnostic Explanations images generated from deep learning-based nasopalatine duct cyst (NPDC) detection models. (A,B) The cropped panoramic radiographs that were true negative in the deep learning-based NPDC detection models. (C,D) The cropped panoramic radiographs that were true positive in the deep learning-based NPDC detection models. Grad-CAM gradient-weighted class activation map**, LIME locally interpretable model-agnostic explanations.
Figure 5 shows cases that were miscategorized by both LeNet and VGG16. There was one case in which non-NPDC was categorized as an NPDC by both LeNet and VGG16. In this case, the ROI included the air space and the hard palate as obstructive shadows. There were three cases in which an NPDC was categorized as non-NPDC by both LeNet and VGG16. In these cases, the ROI included the air space as an obstructive shadow, or the NPDCs were outside of the tomographic layers and had low X-ray transparency on the panoramic radiographs. VGG16 focused along the line of air-containing cavities.
Cropped panoramic radiographs of false negative or false positive cases in the deep learning-based nasopalatine duct cyst (NPDC) detection models, and their corresponding Gradient-weighted Class Activation Map** and Locally Interpretable Model-agnostic Explanations images. (A) The cropped images that were the false positive case in the deep learning-based NPDC detection models. (B–D) The cropped images that were false negative cases in the deep learning-based NPDC detection models. Grad-CAM gradient-weighted class activation map**, LIME locally interpretable model-agnostic explanations.
Discussion
In this study, we performed deep learning of artificial intelligence models that detect NPDCs from panoramic radiographs using a graphical user interface-based convolutional neural network. In this research, preliminary experiments were conducted with many learning settings, made to maximize the accuracy rate. Moreover, deep learning was performed using simple LeNet and VGG16 models to implement the convolutional neural network. Deep learning networks LeNet and VGG16 both had a higher accuracy rate than the radiologists.
LeNet was the first convolutional neural network, proposed by Yann et al. and is characterized by repeating the convolutional layer and the pooling layer twice23. LeNet did not show any obvious overfitting, but the validation error did not converge to the optimal solution. The number of training data should be increased to avoid being trapped in the local minima. Typically, there is also a method to reduce the learning rate, but due to preliminary experiments, a learning rate of 0.001 was optimal. Increasing the batch size reduces the possibility of trap** in the local minima, but due to the number of original data, it was not possible to increase the batch size any further.
The VGG model won second place in the image classification category at the ImageNet Large Scale Visual Recognition Challenge 2014 The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778 (IEEE, Las Vegas, NV, USA, 2016). https://doi.org/10.1109/CVPR.2016.90. Hubel, D. H. & Wiesel, T. N. Receptive fields and functional architecture of monkey striate cortex. J. Physiol. 195, 215–243 (1968). Dhillon, A. & Verma, G. K. Convolutional neural network: A review of models, methodologies and applications to object detection. Prog. Artif. Intell. 9, 85–112 (2020). Chartrand, G. et al. Deep learning: A primer for radiologists. Radiogr. Rev. Publ. Radiol. Soc. N. Ama. 37, 2113–2131 (2017). Zaharchuk, G., Gong, E., Wintermark, M., Rubin, D. & Langlotz, C. P. Deep learning in neuroradiology. AJNR Am. J. Neuroradiol. 39, 1776–1784 (2018). Yasaka, K., Akai, H., Kunimatsu, A., Kiryu, S. & Abe, O. Deep learning with convolutional neural network in radiology. Jpn. J. Radiol. 36, 257–272 (2018). Soffer, S. et al. Convolutional neural networks for radiologic images: A radiologist’s guide. Radiology 290, 590–606 (2019). Small, J. E., Osler, P., Paul, A. B. & Kunst, M. CT cervical spine fracture detection using a convolutional neural network. AJNR Am. J. Neuroradiol. 42, 1341–1347 (2021). Kim, G. R. et al. Convolutional neural network to stratify the malignancy risk of thyroid nodules: Diagnostic performance compared with the American College of Radiology thyroid imaging reporting and data system implemented by experienced radiologists. AJNR Am. J. Neuroradiol. 42, 1513–1519 (2021). Heo, M.-S. et al. Artificial intelligence in oral and maxillofacial radiology: What is currently possible?. Dentomaxillofac. Radiol. 50, 20200375 (2021). Prados-Privado, M., García Villalón, J., Blázquez Torres, A., Martínez-Martínez, C. H. & Ivorra, C. A convolutional neural network for automatic tooth numbering in panoramic images. BioMed. Res. Int. 2021, 3625386 (2021). Schwendicke, F., Golla, T., Dreher, M. & Krois, J. Convolutional neural networks for dental image diagnostics: A sco** review. J. Dent. 91, 103226 (2019). Lee, J.-H., Han, S.-S., Kim, Y. H., Lee, C. & Kim, I. Application of a fully deep convolutional neural network to the automation of tooth segmentation on panoramic radiographs. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 129, 635–642 (2020). Lee, J.-S. et al. Osteoporosis detection in panoramic radiographs using a deep convolutional neural network-based computer-assisted diagnosis system: A preliminary study. Dentomaxillofac. Radiol. 48, 20170344 (2019). Kim, J., Lee, H.-S., Song, I.-S. & Jung, K.-H. DeNTNet: Deep neural transfer network for the detection of periodontal bone loss using panoramic dental radiographs. Sci. Rep. 9, 17615 (2019). Katsumata, A. Deep learning and artificial intelligence in dental diagnostic imaging. Jpn. Dent. Sci. Rev. 59, 329–333 (2023). Ludlow, J. B. & Mol, A. Imaging/intraoral anatomy. In Oral Radiology Principles and Interpretation (eds White, S. C. & Pharoah, M. J.) 138–139 (Mosby, 2014). Koenig, L. J. Cysts, nonodontogenic/mandible and maxilla. In Diagnostic Imaging Oral and Maxillofacial (eds Koenig, L. J. et al.) 66–69 (AMIRSYS, 2012). Vasconcelos, R., de Aguiar, M. F., Castro, W., de Araújo, V. C. & Mesquita, R. Retrospective analysis of 31 cases of nasopalatine duct cyst. Oral Dis. 5, 325–328 (1999). Takagi, R., Ohashi, Y. & Suzuki, M. Squamous cell carcinoma in the maxilla probably originating from a nasopalatine duct cyst: Report of case. J. Oral Maxillofac. Surg. Off. J. Am. Assoc. Oral Maxillofac. Surg. 54, 112–115 (1996). Lee, H.-S. et al. Automatic detection and classification of nasopalatine duct cyst and periapical cyst on panoramic radiographs using deep convolutional neural networks. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 10, 10. https://doi.org/10.1016/j.oooo.2023.09.012 (2023). Ito, K. et al. Characteristic image findings of the nasopalatine duct region using multidetector-row CT. J. Hard Tissue Biol. 25, 69–74 (2016). Lecun, Y., Bottou, L., Bengio, Y. & Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998). Simonyan, K. & Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Preprint at https://doi.org/10.48550/ar**v.1409.1556 (2015). We would like to thank Editage (www.editage.com) for English language editing. This research was supported by JSPS KAKENHI Grant Number JP23K16135. This research was supported by JSPS KAKENHI Grant Number JP23K16135. K.I. contributed to conception, design, data acquisition, analyses, interpretation, and drafted manuscript; N.H. contributed to conception, and critically revised the manuscript; H.M. contributed to design, and critically revised the manuscript; E.S. contributed to interpretation, and critically revised the manuscript; S.T. contributed to data acquisition, and critically revised the manuscript; T.K. contributed to data acquisition, and critically revised the manuscript; T.K. contributed to conception, design, and critically revised the manuscript. All authors gave their final approval and agree to be accountable for all aspects of the work. The authors declare no competing interests. Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Ito, K., Hirahara, N., Muraoka, H. et al. Graphical user interface-based convolutional neural network models for detecting nasopalatine duct cysts using panoramic radiography.
Sci Rep 14, 7699 (2024). https://doi.org/10.1038/s41598-024-57632-8 Received: Accepted: Published: DOI: https://doi.org/10.1038/s41598-024-57632-8Data availability
References
Acknowledgements
Funding
Author information
Authors and Affiliations
Contributions
Corresponding author
Ethics declarations
Competing interests
Additional information
Publisher's note
Rights and permissions
About this article
Cite this article