Abstract
CircRNAs play crucial roles in various malignancies via an increasing number of reported regulatory mechanisms, including the classic sponging mechanism between circRNAs and micro RNAs (miRNAs). We performed bioinformatic analyses and identified circTLK1 as a regulator of malignant chordoma progression. Moreover, we observed that circTLK1 showed high expression in chordoma cells and tissues, while circTLK1 interference suppressed chordoma cell proliferation and invasion. In addition, circTLK1 directly interacted with miR-16-5p, which has previously been shown to repress chordoma, and circTLK1 knockdown suppressed Smad3 expression. Chromatin immunoprecipitation sequencing further demonstrated that Smad3 acts as a positive regulator by interacting with TLK1, thereby mediating the circTLK1/miR-16-5p/Smad3 positive feedback axis. Taken together, our findings suggested that the disruption of the circTLK1/miR-16-5p/Smad3 positive feedback pathway, particularly via the Smad3 inhibitor SIS3, could be a promising therapeutic strategy.
Similar content being viewed by others
Introduction
Chordomas are predominantly located in the spine and cranial basement [1, 2]. Despite having a low incidence (approximately 0.8 patients of one million individuals), chordomas have an alarmingly high rate of recurrence [3] and often do not respond to conventional chemotherapy, which is problematic for researchers and clinical physicians [4, 5]. Consequently, complete surgical resection combined with radiotherapy remain the preferred standard treatment approach [5]. However, a few studies have investigated the mechanisms of initiation, progression, and drug resistance in chordomas.
To date, the molecular pathogenesis of chordomas have been reported to involve impaired brachyury (TBXT) expression, other aberrant genes, and non-coding RNAs (ncRNAs) [6, 7]. However, the study about circRNAs in the chordoma is relatively scarce. Specially, circRNAs are a new type of ncRNAs that were originally thought to be the “garbage byproducts” of precursor RNA splicing [8]. A variety of detected circRNAs exert crucial effects via sponging regulation, protein translation, and transcriptional regulation [9, 10]. For instance, circRNAs can sponge miRNAs to regulate the initiation or proliferation of malignancies such as colorectal cancer, breast cancer, pancreatic ductal adenocarcinoma, and osteosarcoma [11,12,13,14]. In particular, the circRNA CCDC66 was found to promote colon cancer [15], while circPRKCI can promote the growth of lung adenocarcinoma [16] and circ0008399 can promote the m6A methyltransferase complex assembly and activity in bladder cancer [17].
MiRNAs are one of the most important downstream targets that can be sponged by circRNAs. Given that miRNAs interact with the 3′-untranslated region of mRNAs, known as the seed sequence, to suppress their translation, regulatory networks involving circRNAs, miRNAs, and mRNAs could exert considerable effects in various diseases, including malignant cancers [18, 19]. For instance, the miR-181a regulating SFRP4 has been shown in ovarian cancer [20]. Previously, we found that miR-16-5p inhibited chordoma growth and progression via suppressing Smad3 expression [21]. Therefore, having identified circTLK1 which sponge miR-16-5p using starBase v3.0, we further explored its oncogenic effects in chordomas and demonstrated that circTLK1 mediates a positive feedback loop.
Results
Identification of circTLK1 and its features in chordoma
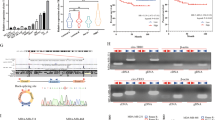
First, we performed bioinformatic analyses based on targetSites, bioComplex and clipReadNum criterion in the starBase v3.0 database and identified three circRNAs targeting miR-16-5p, namely, circTLK1, circCBX4, and circRNMT (Fig. 1A). RT-qPCR showed that circTLK1 was upregulated to a greater degree than other circRNAs in chordoma cells (Fig. 1B).
A Schematic drawing of circTLK1, circCBX4, and circRNMT screened via Starbase. B The relative expression levels of all circRNAs identified in chordoma cells were quantified by RT-qPCR. C The circular structure of circTLK1 was illustrated and the junction site was confirmed by Sanger sequencing. D The agarose gel electrophoresis result showed circTLK1 amplification from cDNA and not gDNA. E The stability of circTLK1 was verified via actinomycin D treatment. F The cellular location of circTLK1 was confirmed by nuclear and cytoplasmic RNA extraction combing with RT-qPCR. G RNA-FISH illustrated the cellular location of circTLK1. H The expression of circTLK1 in chordoma tissues (n = 15) vs pulposus (n = 10) was quantified via RT-qPCR. *P < 0.05, **P < 0.01, ns indicates no significance.
CircTLK1 (circBase ID: hsa_circ_0057021) is composed of exons 11–21 of TLK1 by post-transcriptional back-splicing. Further, we verified the junction site of circTLK1 using Sanger sequencing (Fig. 1C). Since the circular structure may be produced via back-splicing or gene rearrangement, we attempted to confirm its most probable origin by performing RT-qPCR with divergent and convergent primers on cDNA and gDNA extracted from U-CH1 cells. Interestingly, these results showed that circTLK1 was only present in cDNA, and not in gDNA (Fig. 1D). Furthermore, we confirmed that circTLK1 was relatively stable than TLK1 mRNA after actinomycin D treatment (Fig. 1E). The quantity of cytoplasmic RNA demonstrated that circTLK1 existed significantly in the cytoplasm but not nuclei (Fig. 1F). The cellular location of circTLK1 was further confirmed via RNA-fluorescence in situ hybridization (FISH), which showed a dramatic cytoplasmic orientation (Fig. 1G). CircTLK1 was also highly expressed in chordoma tissues (Fig. 1H). Taken together, these findings confirmed the identical characteristics of circTLK1 and its high expression level in chordoma.
CircTLK1 promotes the malignancy of chordoma cell in vitro
Loss-of-function assays were performed by constructing circTLK1-knockout vector. RT-qPCR analysis confirmed that the lentiviral vectors containing sh-circTLK1 substantially interfered with circTLK1 in chordoma cells (Fig. 2A). Notably, circTLK1 interference suppressed chordoma cells proliferation, as confirmed using CCK8 (Fig. 2B), and 5-ethynyl-2′-deoxyuridine (EdU) assay (Fig. 2C). CircTLK1 interference reduced the ability of mobility and invasion (Fig. 2D), as further confirmed by scratch assay (Fig. 2E).
A The efficacy of transfection was verified RT-qPCR. B CCK8 results showed the viability of U-CH1 and MUG-Chor1 cells interfered by sh-circTLK1. C The ability of proliferation was confirmed by EdU assay after silencing circTLK1. D Migration and invasion ability were assessed by Transwell assays after silencing circTLK1. E Wound-healing assay result showed the migration of chordoma cells with circTLK1 interference. F Western blotting results showed the expression level of E-cadherin, N-cadherin, and vimentin proteins with knocked out circTLK1. G CCK8 results showed the decrease of MUG-Chor1 cell viability after transfecting another siRNA. H The mobility of MUG-Chor1 cell transfected with si-circTLK1 was confirmed by wound-healing assay. *P < 0.05, **P < 0.01.
To further elucidate the relationship between circTLK1 and epithelial-mesenchymal transition (EMT), we performed western blotting analysis, which showed that circTLK1 interference promoted E-cadherin, but suppressed N-cadherin and vimentin (Fig. 2F). In addition, to avoid the off-target effects on circTLK1 interference, another si-circTLK1 was involved. The proliferation of chordoma cells was then inhibited after interfering circTLK1 (Fig. 2G). The motility of chordoma cells was inhibited after interfering circTLK1(Fig. 2H). In conclusion, above results suggest that circTLK1 advances the malignancy of chordoma cells.
MiR-16-5p suppresses chordoma malignancy via interacting with Smad3
Based on our previous findings, next, miR-16-5p was interfered and overexpressed. RT-qPCR results showed that the expression level increased in the overexpression group but that of interference group was opposite (Fig. 3A). Subsequent CCK8 demonstrated that chordoma cell viability was reduced by the mimics but promoted by the inhibitors (Fig. 3B).
A The expression level of miR-16-5p was quantified by RT-qPCR in chordoma cells with a miR-16-5p mimic or inhibitor. B CCK8 results indicated the viability in the miR-16-5p mimic or inhibitor group. C, D The expression levels of E-cadherin, N-cadherin, and vimentin proteins were quantified using western blotting. E The interaction between miR-16-5p and Smad3 was verified by a dual-luciferase reporter gene assay. F The expression level of Smad3 protein was quantified by western blot. *P < 0.05, **P < 0.01, ns indicates no significance.
Since EMT progression is of vital importance in chordoma malignancy, related proteins were quantified. All results showed that N-cadherin and vimentin expression deceased but E-cadherin expression increased in overexpression group (Fig. 3C), whereas the interference group exerted the opposite effect (Fig. 3D). The relative intensity of luciferase decreased in wild-type vector (Fig. 3E). Smad3 expression was inhibited by the mimics but promoted by the inhibitors (Fig. 3F), confirming that miR-16-5p interacts with Smad3 to suppress chordoma cell malignancy.
CircTLK1 interacts with miR-16-5p to enhance Smad3 expression
Based on FISH findings of circTLK1 and miR-16-5p, we observed that both predominantly located in the chordoma cellular cytoplasm (Fig. 4A). Luciferase findings revealed the deletion of relative luciferase intensity in the circTLK1-WT group (Fig. 4B). RNA pull down assay indicated that circTLK1 was enriched when biotinylated bead complexes were pulled down (Fig. 4C). Further, proteins combining with circTLK1/miR-16-5p were eluted from pulled down biotinylated bead complexes. The specific interacted protein was identified via silver staining combined with mass spectrum. The results showed that AGO2 protein was the specific functional protein as a mediator between circTLK1 and miR-16-5p (Fig. 4D, E). Western blotting also demonstrated the increased AGO2 protein expression in the biotin-circTLK1 group (Fig. 4F). Next, we used anti-AGO2 antibody to immunoprecipitate the complex produced by RNA immunoprecipitation (RIP) assay, combined with agarose gel electrophoresis illustrating that circTLK1 enrichment was significantly much more in the miR-16-5p mimic group (Fig. 4G). Based on these results, circTLK1/miR-16-5p/Smad3 axis was further verified by western blot, which showed a significantly decreased Smad3 expression level in the sh-circTLK1 group (Fig. 4H). Thus, circTLK1 targets with miR-16-5p to regulate Smad3 and exert oncogenic effects.
A FISH images showed the cellular location of circTLK1 and miR-16-5p. B The target site of the combination was confirmed by a dual-luciferase reporter gene assay. C The interaction between circTLK1 and miR-16-5p was verified using RNA pull-down assay. D Silver staining demonstrated the difference of target proteins between biotin-circTLK1 and biotin-NC. E The Venn diagram showed that specific protein between circTLK1 and miR-16-5p were identified by mass spectrum combining with starBase prediction. F AGO2 protein was assessed by western blot between biotin-circTLK1 and biotin-NC. G RIP assay illustrated the interaction between circTLK1 and miR-16-5p. H The expression level of Smad3 was verified by western blot after transfecting sh-circTLK1. **P < 0.01, ns indicates no significance.
Smad3 interacts with TLK1 to regulate circTLK1 transcription via a positive feedback loop
To clarify the target of Smad3, chromatin immunoprecipitation sequencing (ChIP-seq) was involved. The schematic diagram showed that there are 5.22% DNA fractions located in the promoter region of genes bound by Smad3 (Fig. 5A). The number of the peaks located in promoters with over 6-fold enrichment (p < 1e−20, FDR < 0.01) distributed on every chromosome were counted. It shown that Smad3 combined with chromosomes 1, 2, and 3 with the highest combination (Fig. 5B). There were three peaks observed in the TLK1 gene region of all 20, 371 sequencing results (p < 1e−20, FDR < 0.01; Table 1). Based on the ChIP-seq results, three peaks in the upstream and promoter regions of TLK1 with >6.5-fold enrichment were likely to be Smad3 binding sites (Fig. 5C–E). What’s more, the expression of Smad3 and circTLK1 decreased in chordoma cells after transfecting with si-Smad3#1 and si-Smad3#2 (Fig. 5F). In conclusion, Smad3 functions as a positive modulator that promotes the transcription of circTLK1 through a positive feedback pathway.
CircTLK1 knockdown represses tumor growth
Finally, we confirmed the effect of circTLK1 by injecting BALB/c nude mice with U-CH1 cells to induce subcutaneous tumors (Fig. 6A, B). They showed decreased tumor mass and volume (Fig. 6C–E). H&E staining of tumors was depicted in Fig. 6F, while Smad3 expression was confirmed using IHC in tumors from sh-circTLK1 or sh-NC tumors (Fig. 6G). Furthermore, we observed decreased EMT progression in sh-circTLK1 tumors (Fig. 6H). Therefore, circTLK1 knockdown suppresses chondroma growth in vivo.
A, B The photographs of mice tumors are shown. C–E The volume and weight of the tumors were analyzed and illustrated. F, H & E staining illustrated the pathological characteristics of tumors. G Representative image demonstrated Smad3 expression assessed using IHC. H Expressions of the proteins E-cadherin, N-cadherin, and vimentin were evaluated using western blotting. *P < 0.05, **P < 0.01.
Discussion
CircRNAs participate in various malignant tumors [22,23,24]. However, the quantity of circRNA obtained in chordomas has been low. Previously, we identified miR-16-5p using miRNA microarrays combined with bioinformatic techniques [21] and predicted circTLK1 to be an upstream target of miR-16-5p using starBase [25]. Traditionally the endogenous ceRNA network, the circRNA-miRNA-mRNA axis, has been explored in diverse scopes, including chemotherapy resistance [26], prognostic biomarker [27], malignancy progression [28], and so on. However, in the present study, we explored the novel mechanism through which circTLK1 promotes chordoma progression by regulating a positive feedback pathway involving miR-16-5p/Smad3 (Fig. 7).
Several diagrams are used to indicate the cellular structure and molecules in our study, such as DNA, mRNA, circRNA, and miRNA. The green arrow (↓) indicates the decreased level, conversely, the red one (↑) is the opposite. The interaction is demonstrated by “→” or “⊣”, which means promotion or suppression, respectively.
Dysregulated circRNAs sometimes result from mutant genes and oncogenic viruses like EBV [23]. Regretfully, we were unable to elucidate the underlying aberrant molecular mechanism of this circRNA in chordoma. Aberrant brachyury is considered as one of the initial factors in chordomas [6], but correlation between brachyury and circTLK1 is not investigated in this study. Therefore, although cell function assays demonstrated that aberrant circTLK1 expression was closely associated with chordoma malignancy, the potential mechanism requires further investigation.
Smad3 is a conventional target molecule that has been studied extensively and can be suppressed by miR-16-5p, and that also positively regulates TLK1 transcription. As a positive mediator, Smad3 bridges the circTLK1/miR-16-5p/Smad3 positive feedback pathway. Moreover, Smad3 has been reported to act as a transcription factor that interacts with several genes [29]. Our findings also revealed that Smad3 integrated with the TLK1 gene, acting as a transcription factor. Herein, we believe that the Smad3 inhibitor, SIS3, could be used to attenuate the progression of malignant tumors [30]. SIS3 is a kind of selective Smad3 phosphorylation inhibitor. It has been shown to inhibit the phosphorylation of TGF-β/Smad3 in other cancers, such as colorectal cancer [31]. So, it is increasingly significant to verify the effect and value of SIS3 in chordomas. Regrettably, its therapeutic efficacy in chordoma was not involved in the present study warranting further investigation.
CircRNAs were investigated in our study, nevertheless, its clinical significance remains limited. Avenues to disrupt the positive feedback pathway could be explored further. In addition, future studies should aim to elucidate the molecular mechanism underlying chemotherapy resistance to improve the clinical treatment of chondromas. In conclusion, we demonstrated that circTLK1 dysregulation significantly promotes chordoma progression via a positive feedback loop.
Materials and methods
Human specimens and cell lines
Chordoma (n = 15) and pulposus tissue samples (n = 10) were obtained from patients who underwent surgical resection but no chemotherapy in our institution. All of patients have signed informed consent. This study was approved by the Ethics Committee. Chordoma U-CH1 cell line (Cat# CRL-3217) was purchased from ATCC, and MUG-Chor1 initially available at our laboratory was verified via short tandem repeat analysis. Culture flasks were treated according to the instruction of ATCC. Complete Iscove’s modified Dulbecco’s medium (IMDM; ATCC, Cat# 30-2005) and RPMI-1640 (ATCC, Cat# 30-2001; v/v, 4:1) were used.
Isolation of total RNA and gDNA
Total RNA from chordoma cells (U-CH1 or MUG-Chor1) and tissues frozen in liquid nitrogen was isolated using the RNA extraction kit (Qiagen, Germantown, MD, USA, Cat# 74134) and TRIzol reagent. The DNA micro kit (Qiagen, Cat# 56304) was applied to isolate gDNA. The concentration of the extracted RNA was measured using NanoPhotometer Pearl (IMPLEN, Schatzbogen, Munchen, Germany, Model# 03384).
Nuclear and cytoplasmic RNA extraction
The PARIS™ Kit (Thermo Scientific, Rockford, lL, USA, Cat# AM1921) was used to isolate nuclear and cytoplasmic components. An adequate number of chordoma cells (~106) were lysed in cell fractionation buffer for 5 min. After centrifugation for 3 min, the final nuclear and cytoplasmic RNA were separated and obtained.
RT-qPCR
All premiers were designed, and RT-qPCR was performed to amplify the target genes. CFX96™ System (Bio-Rad) was used. Intrinsic control such as GAPDH as well as U6, was used. All primers’ information is presented in Table 2.
Agarose gel electrophoresis
The separated circRNAs were confirmed using 2% agarose gel electrophoresis (Shanghai Baygene Biotechnology Company Limited, Shanghai, China, Cat# BY-R0100) and the head-to-tail site of circTLK1 was authenticated via Sanger sequencing with RT-qPCR.
Actinomycin D treatment
All cells were treated using 100 ng/mL actinomycin D (Merck, Darmstadt, Germany, Cat# SBR00013) at 0, 6, 12, 18, and 24 h. Thereafter, cells were collected to extract total RNA, and the expression of circTLK1 as well as TLK1 mRNA were quantified via RT-qPCR.
Oligonucleotide, lentivirus, and plasmid transfection
SiRNAs targeting circTLK1 and Smad3 (si-Smad3 #1 and #2), miR-16-5p mimics, and inhibitors were constructed and used to construct sh-circTLK1 or sh-NC lentiviral vectors (Hanbio Biotechnology Company, Shanghai, China). Smad3-overexpression plasmids were purchased from Vigene Biosciences (Shandong, China). All of the cells were transfected with lentivirus supplemented with 0.2% puromycin (Hanbio Biotechnology, Cat# HB-PU-500) for 72 h. Transfection was facilitated with Opti-MEM (Gibco, Cat# 11058021) and Lipofectamine 3000 (Invitrogen, Cat# L3000-015). All the sequences of interference, overexpression, and negative control are listed in Table 3.
Cell proliferation assays
Briefly, 100 μL of complete IMDM medium and 10 μL of CCK8 reagent (Do**do Crop, Japan, Cat# CK04) were added to culture plates containing 8 × 103 cells/well. Data were collected over 4 days serially. Chordoma cells were analyzed using an EdU Kit (Beyotime Biotechnology, Bei**g, China, Cat# C0075S) to confirm cell proliferation.
Wound-healing
Chordoma cells were prepared (1 × 106/well) overnight and an artificial wound was scratched. The photographs at the initial and three days were taken under a microscope (Leica Microsystems Inc, Bannockburn, IL, USA), then the width of the wound was measured and analyzed.
Transwell migration and invasion assays
Transwell chambers (Corning, NY, USA, Cat# 3422) with an 8-μm pore size were used. Specially, 300 μL FBS-free IMDM medium containing 5 × 104 chordoma cells was added up to the top of the chamber. After culturing for 2 days, the following steps were referred to the relevant article [21, 26]. Photographs were taken under the microscope (Leica Microsystems Inc), and migratory cells were counted as well as analyzed.
Western blotting
Equivalent quantity of protein was added up to 10% SDS-PAGE (Epizyme Biomedical Technology, Shanghai, China, Cat# PG112). Then transferred PVDF membranes (Merk Millipore, Tulagreen, Carrigtwohill, Co Cork IRELAND, Cat# IPVH00010) were blocked with 1× fast-blocking liquid, then incubated with antibodies against Smad3 (1:1000; Cell Signaling Technology, Cat# 9523S), E-cadherin (1:5000, Proteintech, Wuhan, China, Cat# 20874-1-AP), N-cadherin (1:5000; Proteintech, Cat# 22018-1-AP), vimentin (1:5000; Proteintech, Cat# 10366-1-AP), and GAPDH (1:5000; Proteintech, Cat# 10494-1-AP). Specific horseradish peroxidase (HRP)-conjugated secondary antibodies against IgG-mouse or rabbit were hybridized with the primary antibodies in the 37 °C incubator.
FISH
Firstly, U-CH1 and MUG-Chor1 cells slides were prepared. Then circTLK1 probe and miR-16-5p probe were designed. During preparation, all slides were prepared with being fixed. For pre-hybridization, the slides were incubated at 37 °C. The following steps, hybridization-washing-hybridization, were carried out. Finally, the photographs were taken under a scanning confocal microscope (Leica Microsystems Inc).
Dual-luciferase gene reporter assay
Both wild and mutant-type vectors were constructed in GenePharma company. In brief, MUG-Chor1 cells transfected with wild and mutant-type circTLK1 or Smad3 were lysed followed by a dual-luciferase assay kit (Beyotime Biotechnology, Cat# RG027). Finally, a microplate reader (Bio-Tek, Santa Clara, CA, USA, Model No. SLXFA) was used to detect relative luciferase intensity.
IHC staining
Briefly, after deparaffinization, the sections were incubated at 4 °C overnight with corresponding antibodies. The following process was according to the IHC kit (Boster Biological Technology, China, Cat# SA2010). The concentration of Smad3 antibody was used with 1:800. The sections were observed and photos were taken using a Leica microscope.
RIP
The lysate of MUG-Chor1 cells (~2 × 107) had been prepared with using RIP lysis buffer of EZ-Magna Protein Immunoprecipitation Kit (Millipore, Cat# 17-701), and the lysate were bound to beads-Argonaute 2 (Proteintech, Cat# 10686-1-AP) or IgG antibody complex via gentle agitation overnight at 4 °C.
RNA pull-down assay
Biotinylated circTLK1 and miR-16-5p were constructed initially, and RNA pull-down assay was performed using Pierce Pull-Down Kit (Thermo Scientific, Cat# 20164). The lysate was agitated gently with M-270 magnetic beads overnight. CircTLK1 and miR-16-5p pull-down were quantified using RT-qPCR. In addition, RNA binding proteins were also purified to identify the specific protein between circTLK1 and miR-16-5p using MS.
ChIP-seq
A ChIP Kit (Millipore, Cat# 17-610) was applied to chromatin immunoprecipitation (ChIP) assays. In brief, MUG-Chor1 were cross-linked and lysed using lysis buffer. Cellular chromatin was disrupted using ultrasound and then immunoprecipitated using anti-Smad3 antibodies. The target DNA fractions were sequenced and analyzed.
In vivo animal experiment
The animal experiment was approved by Ethics Committee of our institute. To confirm the oncogenic effect of circTLK1, BALB/c nude mice (n = 5 per group) purchased from Bei**g Viltalriver were subcutaneously injected with 5 × 106 U-CH1 cells. All mice were separated into different two group at random and blindly. Tumor volume was recorded every week. All mice had been euthanized with the tumor volume not exceeding 2 cm3.
Statistical analysis
Statistical analyses were carried out using SPSS software (v22.0; Armonk, NY, USA) and GraphPad Prism 9.0.0. The method of Student’s t-test was involved. No significance was indicated by ns. Significant data are indicated by *P < 0.05, **P < 0.01, ***P < 0.001.
Data availability
The data that were collected and analyzed during the current study are available from the corresponding author on reasonable request.
References
Barber SM, Sadrameli SS, Lee JJ, Fridley JS, Teh BS, Oyelese AA, et al. Chordoma-current understanding and modern treatment paradigms. J Clin Med. 2021;10:1054.
Levine AB, Wong D, Fatehi M, Yip S. Ependymoma and chordoma. Neurosurgery. 2020;87:860–70.
Ulici V, Hart J. Chordoma. Arch Pathol Lab Med. 2022;146:386–95. https://doi.org/10.5858/arpa.2020-0258-RA
Karpathiou G, Dumollard JM, Dridi M, Dal Col P, Barral FG, Boutonnat J, et al. Chordomas: a review with emphasis on their pathophysiology, pathology, molecular biology, and genetics. Pathol Res Pr. 2020;216:153089.
Gulluoglu S, Turksoy O, Kuskucu A, Ture U, Bayrak OF. The molecular aspects of chordoma. Neurosurg Rev. 2016;39:185–96.
Cottone L, Cribbs AP, Khandelwal G, Wells G, Ligammari L, Philpott M, et al. Inhibition of histone H3K27 demethylases inactivates brachyury (TBXT) and promotes chordoma cell death. Cancer Res. 2020;80:4540–51.
Kitamura Y, Sasaki H, Yoshida K. Genetic aberrations and molecular biology of skull base chordoma and chondrosarcoma. Brain Tumor Pathol. 2017;34:78–90.
Huang Y, Zhang C, **ong J, Ren H. Emerging important roles of circRNAs in human cancer and other diseases. Genes Dis. 2021;8:412–23.
Tao M, Zheng M, Xu Y, Ma S, Zhang W, Ju S. CircRNAs and their regulatory roles in cancers. Mol Med. 2021;27:94.
Jiang MP, Xu WX, Hou JC, Xu Q, Wang DD, Tang JH. The emerging role of the interactions between circular RNAs and RNA-binding proteins in common human cancers. J Cancer. 2021;12:5206–19.
Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20:93.
Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B, et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. 2021;20:138.
Rong Z, Shi S, Tan Z, Xu J, Meng Q, Hua J, et al. Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Mol Cancer. 2021;20:106.
Chen J, Liu G, Wu Y, Ma J, Wu H, **e Z, et al. CircMYO10 promotes osteosarcoma progression by regulating miR-370-3p/RUVBL1 axis to enhance the transcriptional activity of β-catenin/LEF1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18:150.
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–50.
Qiu M, **a W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–51.
Wei W, Sun J, Zhang H, **ao X, Huang C, Wang L, et al. Circ0008399 interaction with WTAP promotes assembly and activity of the m6A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res. 2021;81:6142–56.
Mohanapriya R, Akshaya RL, Selvamurugan N. A regulatory role of circRNA-miRNA-mRNA network in osteoblast differentiation. Biochimie. 2022;193:137–47.
He C, Duan L, Zheng H, Li-Ling J, Song L, Li L. Graph convolutional network approach to discovering disease-related circRNA-miRNA-mRNA axes. Methods. 2022;198:45–55.
Belur Nagaraj AB, Knarr M, Sekhar S, Connor RS, Joseph P, Kovalenko O, et al. The miR-181a-SFRP4 axis regulates Wnt activation to drive stemness and platinum resistance in ovarian cancer. Cancer Res. 2021;81:2044–55.
Zhang H, Yang K, Ren T, Huang Y, Tang X, Guo W. miR-16-5p inhibits chordoma cell proliferation, invasion and metastasis by targeting Smad3. Cell Death Dis. 2018;9:680.
Deng G, Wang R, Sun Y, Huang CP, Yeh S, You B, et al. Targeting androgen receptor (AR) with antiandrogen Enzalutamide increases prostate cancer cell invasion yet decreases bladder cancer cell invasion via differentially altering the AR/circRNA-ARC1/miR-125b-2-3p or miR-4736/PPARγ/MMP-9 signals. Cell Death Differ. 2021;28:2145–59.
Ge J, Wang J, **ong F, Jiang X, Zhu K, Wang Y, et al. Epstein-Barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–88.
Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L, et al. circIGHG-induced epithelial-to-mesenchymal transition promotes oral squamous cell carcinoma progression via miR-142-5p/IGF2BP3 signaling. Cancer Res. 2021;81:344–55.
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–7.
Li S, Liu F, Zheng K, Wang W, Qiu E, Pei Y, et al. CircDOCK1 promotes the tumorigenesis and cisplatin resistance of osteogenic sarcoma via the miR-339-3p/IGF1R axis. Mol Cancer. 2021;20:161.
Chen Y, Huang X, Zhu K, Li C, Peng H, Chen L, et al. LIMD2 is a prognostic and predictive marker in patients with esophageal cancer based on a ceRNA network analysis. Front Genet. 2021;12:774432.
Zhao CX, Yan ZX, Wen JJ, Fu D, Xu PP, Wang L, et al. CircEAF2 counteracts Epstein-Barr virus-positive diffuse large B-cell lymphoma progression via miR-BART19-3p/APC/β-catenin axis. Mol Cancer. 2021;20:153.
Yang C, Chen XC, Li ZH, Wu HL, **g KP, Huang XR, et al. SMAD3 promotes autophagy dysregulation by triggering lysosome depletion in tubular epithelial cells in diabetic nephropathy. Autophagy. 2021;17:2325–44.
Zeng D, **ao Z, Xu Q, Luo H, Wen L, Tang C, et al. Norcantharidin protects against renal interstitial fibrosis by suppressing TWEAK-mediated Smad3 phosphorylation. Life Sci. 2020;260:118488.
Hou B, Li W, **a P, Zhao F, Liu Z, Zeng Q, et al. LHPP suppresses colorectal cancer cell migration and invasion in vitro and in vivo by inhibiting Smad3 phosphorylation in the TGF-β pathway. Cell Death Disco. 2021;7:273.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81972509) and the Bei**g Science and Technology Planning Project (Z171100001017097).
Author information
Authors and Affiliations
Contributions
JL designed the experiment and wrote this manuscript. HZ designed the study methods and provided the technical support. QH tackled with data process. CC, WW, and JN were responsible for the manuscript review. JX assisted with collecting clinical specimens. TR and YH provided me an instruction with facilities usage. XT and WG supported financial resource and scheme supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
Relevant procedures in this project were approved by Ethics Committee of our institute at Peking University People’s Hospital.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lou, J., Zhang, H., Huang, Q. et al. The malignancy of chordomas is enhanced via a circTLK1/miR-16-5p/Smad3 positive feedback axis. Cell Death Discov. 9, 64 (2023). https://doi.org/10.1038/s41420-023-01332-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01332-1
- Springer Nature Limited
This article is cited by
-
Dysregulation of noncoding RNA in chordoma; implications in identifying potential targets for novel therapeutic approaches
Molecular Biology Reports (2024)