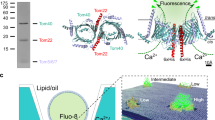
Abstract
Tim23, the central subunit of the TIM23 protein-translocation complex, forms a voltage-gated channel in the mitochondrial inner membrane (MIM), an energy-conserving membrane that generates a proton-motive force to drive vital processes. Using high-resolution fluorescence map** of a channel-facing transmembrane segment (TMS2) of Tim23 from Saccharomyces cerevisiae, we demonstrate that changes in the energized state of the MIM cause marked structural alterations in the channel region. In an energized membrane, TMS2 forms a continuous α-helix that is inaccessible to the aqueous intermembrane space (IMS). Upon depolarization, the helical periodicity of TMS2 is disrupted, and the channel becomes exposed to the IMS. Kinetic measurements confirm that changes in TMS2 conformation coincide with depolarization. These results reveal how the energized state of the membrane drives functionally relevant structural dynamics in membrane proteins coupled to processes such as channel gating.






Similar content being viewed by others
References
Loew, L.M., Tuft, R.A., Carrington, W. & Fay, F.S. Imaging in five dimensions: time-dependent membrane potentials in individual mitochondria. Biophys. J. 65, 2396–2407 (1993).
Wang, L. Measurements and implications of the membrane dipole potential. Annu. Rev. Biochem. 81, 615–635 (2012).
Demchenko, A.P. & Yesylevskyy, S.O. Nanoscopic description of biomembrane electrostatics: results of molecular dynamics simulations and fluorescence probing. Chem. Phys. Lipids 160, 63–84 (2009).
Hille, B. Ion Channels of Excitable Membranes (Sinauer, 2001).
Chacinska, A., Koehler, C.M., Milenkovic, D., Lithgow, T. & Pfanner, N. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 (2009).
Endo, T., Yamano, K. & Kawano, S. Structural insight into the mitochondrial protein import system. Biochim. Biophys. Acta 1808, 955–970 (2011).
Gebert, N., Ryan, M.T., Pfanner, N., Wiedemann, N. & Stojanovski, D. Mitochondrial protein import machineries and lipids: a functional connection. Biochim. Biophys. Acta 1808, 1002–1011 (2011).
Marom, M., Azem, A. & Mokranjac, D. Understanding the molecular mechanism of protein translocation across the mitochondrial inner membrane: still a long way to go. Biochim. Biophys. Acta 1808, 990–1001 (2011).
Alder, N.N. Biogenesis of lipids and proteins within biological membranes. in The Structure of Biological Membranes (ed. Yeagle, P.L.) 315–377 (CRC, 2011).
Rassow, J., Hartl, F.U., Guiard, B., Pfanner, N. & Neupert, W. Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett. 275, 190–194 (1990).
Geissler, A. et al. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell 111, 507–518 (2002).
Mokranjac, D. et al. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 22, 816–825 (2003).
Truscott, K.N. et al. A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8, 1074–1082 (2001).
Yamamoto, H. et al. Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell 111, 519–528 (2002).
Marom, M. et al. Direct interaction of mitochondrial targeting presequences with purified components of the TIM23 protein complex. J. Biol. Chem. 286, 43809–43815 (2011).
Schulz, C. et al. Tim50's presequence receptor domain is essential for signal driven transport across the TIM23 complex. J. Cell Biol. 195, 643–656 (2011).
Gebert, M. et al. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J. Cell Biol. 197, 595–604 (2012).
Chacinska, A. et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 (2005).
Chacinska, A. et al. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell Biol. 30, 307–318 (2010).
Mokranjac, D., Popov-Celeketic, D., Hell, K. & Neupert, W. Role of Tim21 in mitochondrial translocation contact sites. J. Biol. Chem. 280, 23437–23440 (2005).
Tamura, Y. et al. Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141 (2009).
van der Laan, M. et al. A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr. Biol. 16, 2271–2276 (2006).
Wiedemann, N., van der Laan, M., Hutu, D.P., Rehling, P. & Pfanner, N. Sorting switch of mitochondrial presequence translocase involves coupling of motor module to respiratory chain. J. Cell Biol. 179, 1115–1122 (2007).
Glick, B.S. et al. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809–822 (1992).
Botelho, S.C. et al. TIM23-mediated insertion of transmembrane α-helices into the mitochondrial inner membrane. EMBO J. 30, 1003–1011 (2011).
van der Laan, M. et al. Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9, 1152–1159 (2007).
Martin, J., Mahlke, K. & Pfanner, N. Role of an energized inner membrane in mitochondrial protein import: Δψ drives the movement of presequences. J. Biol. Chem. 266, 18051–18057 (1991).
Alder, N.N., Jensen, R.E. & Johnson, A.E. Fluorescence map** of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell 134, 439–450 (2008).
Lohret, T.A., Jensen, R.E. & Kinnally, K.W. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J. Cell Biol. 137, 377–386 (1997).
Martinez-Caballero, S., Grigoriev, S.M., Herrmann, J.M., Campo, M.L. & Kinnally, K.W. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J. Biol. Chem. 282, 3584–3593 (2007).
Bauer, M.F., Sirrenberg, C., Neupert, W. & Brunner, M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87, 33–41 (1996).
Meier, S., Neupert, W. & Herrmann, J.M. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J. Biol. Chem. 280, 7777–7785 (2005).
Meinecke, M. et al. Tim50 maintains the permeability barrier of the mitochondrial inner membrane. Science 312, 1523–1526 (2006).
Huang, S., Ratliff, K.S. & Matouschek, A. Protein unfolding by the mitochondrial membrane potential. Nat. Struct. Biol. 9, 301–307 (2002).
Alder, N.N., Sutherland, J., Buhring, A.I., Jensen, R.E. & Johnson, A.E. Quaternary structure of the mitochondrial TIM23 complex reveals dynamic association between Tim23p and other subunits. Mol. Biol. Cell 19, 159–170 (2008).
Geissler, A. et al. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b2 modulates the Δψ-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell 11, 3977–3991 (2000).
Lancet, D. & Pecht, I. Spectroscopic and immunochemical studies with nitrobenzoxadiazolealanine, a fluorescent dinitrophenyl analogue. Biochemistry 16, 5150–5157 (1977).
Lin, S. & Struve, W.S. Time-resolved fluorescence of nitrobenzoxadiazole-aminohexanoic acid: effect of intermolecular hydrogen-bonding on non-radiative decay. Photochem. Photobiol. 54, 361–365 (1991).
Cornette, J.L. et al. Hydrophobicity scales and computational techniques for detecting amphipathic structures in proteins. J. Mol. Biol. 195, 659–685 (1987).
Silberberg, S.D., Chang, T.H. & Swartz, K.J. Secondary structure and gating rearrangements of transmembrane segments in rat P2X4 receptor channels. J. Gen. Physiol. 125, 347–359 (2005).
Ma, Z., Kong, J. & Kallen, R.G. Studies of α-helicity and intersegmental interactions in voltage-gated Na+ channels: S2D4. PLoS ONE 4, e7674 (2009).
Kim, P.K., Annis, M.G., Dlugosz, P.J., Leber, B. & Andrews, D.W. During apoptosis bcl-2 changes membrane topology at both the endoplasmic reticulum and mitochondria. Mol. Cell 14, 523–529 (2004).
Baracca, A., Sgarbi, G., Solaini, G. & Lenaz, G. Rhodamine 123 as a probe of mitochondrial membrane potential: evaluation of proton flux through F0 during ATP synthesis. Biochim. Biophys. Acta 1606, 137–146 (2003).
Hall, S.E., Roberts, K. & Vaidehi, N. Position of helical kinks in membrane protein crystal structures and the accuracy of computational prediction. J. Mol. Graph. Model. 27, 944–950 (2009).
Langelaan, D.N., Wieczorek, M., Blouin, C. & Rainey, J.K. Improved helix and kink characterization in membrane proteins allows evaluation of kink sequence predictors. J. Chem. Inf. Model. 50, 2213–2220 (2010).
Screpanti, E. & Hunte, C. Discontinuous membrane helices in transport proteins and their correlation with function. J. Struct. Biol. 159, 261–267 (2007).
Chakrapani, S., Sompornpisut, P., Intharathep, P., Roux, B. & Perozo, E. The activated state of a sodium channel voltage sensor in a membrane environment. Proc. Natl. Acad. Sci. USA 107, 5435–5440 (2010).
White, S.H. & Wimley, W.C. Membrane protein folding and stability: physical principles. Annu. Rev. Biophys. Biomol. Struct. 28, 319–365 (1999).
Møller, J.V., Olesen, C., Winther, A.M. & Nissen, P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 43, 501–566 (2010).
Cao, Z. & Bowie, J.U. Shifting hydrogen bonds may produce flexible transmembrane helices. Proc. Natl. Acad. Sci. USA 109, 8121–8126 (2012).
Erlandson, K.J. et al. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature 455, 984–987 (2008).
Swartz, K.J. Sensing voltage across lipid membranes. Nature 456, 891–897 (2008).
Zorov, D.B. et al. Regulation and pharmacology of the mitochondrial permeability transition pore. Cardiovasc. Res. 83, 213–225 (2009).
Hüser, J., Rechenmacher, C.E. & Blatter, L.A. Imaging the permeability pore transition in single mitochondria. Biophys. J. 74, 2129–2137 (1998).
Ly, J.D., Grubb, D.R. & Lawen, A. The mitochondrial membrane potential Δψ(m) in apoptosis; an update. Apoptosis 8, 115–128 (2003).
Hüser, J. & Blatter, L.A. Fluctuations in mitochondrial membrane potential caused by repetitive gating of the permeability transition pore. Biochem. J. 343, 311–317 (1999).
Buckman, J.F. & Reynolds, I.J. Spontaneous changes in mitochondrial membrane potential in cultured neurons. J. Neurosci. 21, 5054–5065 (2001).
O'Reilly, C.M., Fogarty, K.E., Drummond, R.M., Tuft, R.A. & Walsh, J.V. Jr. Quantitative analysis of spontaneous mitochondrial depolarizations. Biophys. J. 85, 3350–3357 (2003).
Schwarzländer, M. et al. Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. Plant Cell 24, 1188–1201 (2012).
Daum, G., Bohni, P.C. & Schatz, G. Import of proteins into mitochondria: cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033 (1982).
Erickson, A.H. & Blobel, G. Cell-free translation of messenger RNA in a wheat germ system. Methods Enzymol. 96, 38–50 (1983).
Krayl, M., Lim, J.H., Martin, F., Guiard, B. & Voos, W. A cooperative action of the ATP-dependent import motor complex and the inner membrane potential drives mitochondrial preprotein import. Mol. Cell. Biol. 27, 411–425 (2007).
Pfanner, N. et al. Energy requirements for unfolding and membrane translocation of precursor proteins during import into mitochondria. J. Biol. Chem. 265, 16324–16329 (1990).
Crowley, K.S., Liao, S., Worrell, V.E., Reinhart, G.D. & Johnson, A.E. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461–471 (1994).
Crowley, K.S., Reinhart, G.D. & Johnson, A.E. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell 73, 1101–1115 (1993).
Acknowledgements
This work was supported by grants from the US National Science Foundation (MCB-1024908 to N.N.A.), the US National Institutes of Health (GM26494 to A.E.J.) and the Robert A. Welch Foundation (Chair Grant BE-0017 to A.E.J.).
Author information
Authors and Affiliations
Contributions
All authors conceived of and designed the research; K.M., M.S. and N.N.A. conducted experiments; all authors analyzed data; N.N.A. supervised the project and prepared the manuscript and supplementary material.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Combined PDF
Supplementary Figures 1–7 and Supplementary Notes 1–3 (PDF 4687 kb)
Rights and permissions
About this article
Cite this article
Malhotra, K., Sathappa, M., Landin, J. et al. Structural changes in the mitochondrial Tim23 channel are coupled to the proton-motive force. Nat Struct Mol Biol 20, 965–972 (2013). https://doi.org/10.1038/nsmb.2613
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2613
- Springer Nature America, Inc.