Abstract
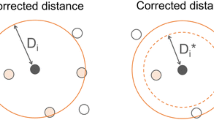
One of the most important features of QSPAR models is their predictive ability. The predictive ability of QSPAR models should be checked by external validation. In this work we examined three different types of external validation set selection methods for their usefulness in in-silico screening. The usefulness of the selection methods was studied in such a way that: 1) We generated thousands of QSPR models and stored them in `model banks'. 2) We selected a final top model from the model banks based on three different validation set selection methods. 3) We predicted large data sets, which we called `chemical universe sets', and calculated the corresponding SEPs. The models were generated from small fractions of the available water solubility data during a GA Variable Subset Selection procedure. The external validation sets were constructed by random selections, uniformly distributed selections or by perimeter-oriented selections. We found that the best performing models on the perimeter-oriented external validation sets usually gave the best validation results when the remaining part of the available data was overwhelmingly large, i.e., when the model had to make a lot of extrapolations. We also compared the top final models obtained from external validation set selection methods in three independent and different sizes of `chemical universe sets'.
Similar content being viewed by others
References
Wermuth, C.-G., Ganellin, C. R., Lindberg, P. and Mitscher, L. A., Glossary of terms used in medicinal chemistry, Pure Appl. Chem., 70 (1998) 1129–1143.
Őrfi, L. and Kövesdi, I., Lead Search, Selection and Optimization in Silico (Virtual) Screening, in Kéri, G. and Tóth, I. (eds.), Molecular Pathomechanisms and New Trends in Drug Research, Taylor & Francis, London, 2003, pp 166–177, ISBN: 0-415-27725-6.
Golbraikh, A. and Tropsha, A., Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection, Mol. Div., 5 (2000) 231–243.
Ran, Y., Jain, N. and Yalkowsky, S. H., Prediction of aqueous solubility of organic compounds by the general solubility equation (GSE), J. Chem. Inf. Comput. Sci., 41 (2001) 1208–1217.
Huuskonen, J., Marja Salo, M. and Taskinen, J., Aqueous solubility prediction of drugs based on molecular topology and neural network modeling, J. Chem. Inf. Comput. Sci., 38 (1998) 450–456.
Mitchell, B. E. and Jurs, P. C., Prediction of aqueous solubility of organic compounds from molecular structure, J. Chem. Inf. Comput. Sci., 38 (1998) 489–496.
McElroy, N. R. and Jurs. P. C., Prediction of aqueous solubility of heteroatom-containing organic compounds from molecular structure., J. Chem. Inf. Comput. Sci., 41 (2001) 1237–1247.
McFarland, J. W., Estimating the water solubilities of crystalline compounds from their chemical structures alone, J.Chem. Inf. Comput. Sci., 41 (2001) 1355–1359.
. Yaffe, D., Cohen, Y., Espinosa, G., Arenas, A. and Giralt, F., A fuzzy ARTMAP based on quantitative structure-property relationships (QSPRs) for predicting aqueous solubility of organic compounds, J. Chem. Inf. Comput. Sci., 41 (2001) 1177–1207.
Huuskonen, J., Estimation of aqueous solubility for diverse set of organic compounds based on molecular topology, J.Chem. Inf. Comput. Sci., 40 (2000) 773–777.
DRAGON Web version 3.0, Talete srl., 1997–2003.
Snee, R. D., Validation of regression-models-methods and examples, Technometrics, 19 (1977) 415–428.
Johnson, M. E., Ylvisaker, D. and Moore, L., Minimax and maximin distance designs, J. Stat. Plan. Infer., 26 (1990) 131–148.
Erös, D., Kéri G., Kövesdi I., Szántai-Kis C., Mészáros G. and Őrfi L., Comparison of predictive ability of water solubility QSPR models generated by MLR, PLS and ANN methods, in press, Mini Rew. in. Med. Chem. (2003).
Erös D., Kövesdi I., Őrfi L., Takács-Novák K., Acsády G. and Kéri G., Reliability of logP predictions based on calculated molecular descriptors: A critical review, Curr. Med. Chem. 9 (2002) 1819–1829.
3-DNET4W 1.1.36, Vichem Chemie Ltd., 1022 Budapest, Herman O u. 15, 2001–2003.
Kövesdi, I., Kéri, G. and Őrfi, L., Method for Generating a Quantitative Structure Property Activity Relationship, WO 02/082329 (2001).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Szántai-Kis, C., Kövesdi, I., Kéri, G. et al. Validation subset selections for extrapolation oriented QSPAR models. Mol Divers 7, 37–43 (2003). https://doi.org/10.1023/B:MODI.0000006538.99122.00
Issue Date:
DOI: https://doi.org/10.1023/B:MODI.0000006538.99122.00