Abstract
This study delves into the functional intricacies of lipoate ligase A (LplA), an enzyme showing great promise in bioconjugation due to its unique capacity for introducing azido groups into proteins without requiring a genetic tag. We aimed to enhance the understanding of LplA's functionality, particularly its substrate tolerance and the reliability of various analytical techniques. A pivotal aspect of our approach was incorporating azido groups into a range of proteins, followed by the addition of the fluorescent molecule Cy3 via click chemistry. Analysis of fluorescent intensity in the altered proteins indicated varying degrees of conjugation. Additionally, phenyl resin-based RP-HPLC facilitated effective separation of modified proteins, unmodified proteins, and remaining fluorescent tags post-separation. SASA analysis provided insights into conjugation trends, guiding the identification of proteins amenable to LplA's tag-free modification. Our findings demonstrate LplA's broad substrate tolerability for protein modification.
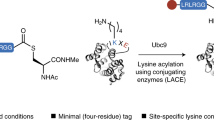
Graphical Abstract




Similar content being viewed by others
Data availability
Not applicable.
References
S. Yamazaki, Y. Matsuda, Tag-free enzymatic modification for antibody−drug conjugate production. ChemistrySelect 7(48), e202203753 (2022). https://doi.org/10.1002/slct.202203753
A.F. Hussain, A. Grimm, W. Sheng, C. Zhang, M. Al-Rawe, K. Brautigam, M. Abu Mraheil, F. Zeppernick, I. Meinhold-Heerlein, Toward homogenous antibody drug conjugates using enzyme-based conjugation approaches. Pharmaceuticals (Basel) 14(4), 343 (2021). https://doi.org/10.3390/ph14040343
A. Beygmoradi, A. Homaei, R. Hemmati, P. Fernandes, Recombinant protein expression: challenges in production and folding related matters. Int. J. Biol. Macromol.Macromol. 233, 123407 (2023). https://doi.org/10.1016/j.ijbiomac.2023.123407
Z. Tawfiq, N.C. Caiazza, S. Kambourakis, Y. Matsuda, B. Griffin, J.C. Lippmeier, B.A. Mendelsohn, Synthesis and biological evaluation of antibody drug conjugates based on an antibody expression system: conamax. ACS Omega 5(13), 7193–7200 (2020). https://doi.org/10.1021/acsomega.9b03628
P.R. Spycher, C.A. Amann, J.E. Wehrmuller, D.R. Hurwitz, O. Kreis, D. Messmer, A. Ritler, A. Kuchler, A. Blanc, M. Behe, P. Walde, R. Schibli, Dual, site-specific modification of antibodies by using solid-phase immobilized microbial transglutaminase. ChemBioChem 18(19), 1923–1927 (2017). https://doi.org/10.1002/cbic.201700188
S. Dickgiesser, M. Rieker, D. Mueller-Pompalla, C. Schroter, J. Tonillo, S. Warszawski, S. Raab-Westphal, S. Kuhn, T. Knehans, D. Konning, J. Dotterweich, U.A.K. Betz, J. Anderl, S. Hecht, N. Rasche, Site-specific conjugation of native antibodies using engineered microbial transglutaminases. Bioconjug. Chem.. Chem. 31(4), 1070–1076 (2020). https://doi.org/10.1021/acs.bioconjchem.0c00061
A. Hadjabdelhafid-Parisien, S. Bitsch, A. Macarron Palacios, L. Deweid, H. Kolmar, J.N. Pelletier, Tag-free, specific conjugation of glycosylated IgG1 antibodies using microbial transglutaminase. RSC Adv. 12(52), 33510–33515 (2022). https://doi.org/10.1039/d2ra05630e
T.W. Morris, K.E. Reed, J.E. Cronan, Identification of the gene encoding lipoate-protein ligase A of Escherichia coli. Molecular cloning and characterization of the lplA gene and gene product. J. Biol. Chem. 269(23), 16091–16100 (1994). https://doi.org/10.1016/s0021-9258(17)33977-7
M. Baalmann, M. Best, R. Wombacher, Site-specific protein labeling utilizing lipoic acid ligase (LplA) and bioorthogonal inverse electron demand Diels-Alder reaction. Methods Mol. Biol. 1728, 365–387 (2018). https://doi.org/10.1007/978-1-4939-7574-7_23
J.G. Plaks, J.L. Kaar, Lipoic acid ligase-promoted bioorthogonal protein modification and immobilization. Methods Mol. Biol. 2012, 279–297 (2019). https://doi.org/10.1007/978-1-4939-9546-2_14
S. Puthenveetil, D.S. Liu, K.A. White, S. Thompson, A.Y. Ting, Yeast display evolution of a kinetically efficient 13-amino acid substrate for lipoic acid ligase. J. Am. Chem. Soc. 131(45), 16430–16438 (2009). https://doi.org/10.1021/ja904596f
S. Yamazaki, N. Shikida, K. Takahashi, Y. Matsuda, K. Inoue, K. Shimbo, Y. Mihara, Lipoate-acid ligase a modification of native antibody: synthesis and conjugation site analysis. Bioorg. Med. Chem. Lett.. Med. Chem. Lett. 51, 128360 (2021). https://doi.org/10.1016/j.bmcl.2021.128360
S. Yamazaki, K. Inoue, Y. Mihara, Y. Matsuda, Tag-free antibody modification mediated by lipoic acid ligase a: application to antibody-drug conjugates production. ChemistrySelect 8(9), e202204706 (2023). https://doi.org/10.1002/slct.202204706
Y. Matsuda, M. Leung, Z. Tawfiq, T. Fujii, B.A. Mendelsohn, In-situ reverse phased HPLC analysis of intact antibody-drug conjugates. Anal. Sci. 37(8), 1171–1176 (2021). https://doi.org/10.2116/analsci.20P424
E. Durham, B. Dorr, N. Woetzel, R. Staritzbichler, J. Meiler, Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J. Mol. Model. 15(9), 1093–1108 (2009). https://doi.org/10.1007/s00894-009-0454-9
T. Fujii, Y. Matsuda, T. Seki, N. Shikida, Y. Iwai, Y. Ooba, K. Takahashi, M. Isokawa, S. Kawaguchi, N. Hatada, T. Watanabe, R. Takasugi, A. Nakayama, K. Shimbo, B.A. Mendelsohn, T. Okuzumi, K. Yamada, AJICAP second generation: improved chemical site-specific conjugation technology for antibody-drug conjugate production. Bioconjug. Chem.. Chem. 34(4), 728–738 (2023). https://doi.org/10.1021/acs.bioconjchem.3c00040
Y. Nakahara, B.A. Mendelsohn, Y. Matsuda, Antibody-drug conjugate synthesis using continuous flow microreactor technology. Org. Process Res. Dev. 26(9), 2766–2770 (2022). https://doi.org/10.1021/acs.oprd.2c00217
P.E. Stein, A.G. Leslie, J.T. Finch, R.W. Carrell, Crystal structure of uncleaved ovalbumin at 1.95 A resolution. J. Mol. Biol. 221(3), 941–959 (1991). https://doi.org/10.1016/0022-2836(91)80185-w
H. Kurokawa, J.C. Dewan, B. Mikami, J.C. Sacchettini, M. Hirose, Crystal structure of hen apo-ovotransferrin. Both lobes adopt an open conformation upon loss of iron. J. Biol. Chem. 274(40), 28445–28452 (1999). https://doi.org/10.1074/jbc.274.40.28445
K.A. Majorek, P.J. Porebski, A. Dayal, M.D. Zimmerman, K. Jablonska, A.J. Stewart, M. Chruszcz, W. Minor, Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol. Immunol.Immunol. 52(3–4), 174–182 (2012). https://doi.org/10.1016/j.molimm.2012.05.011
S.Y. Wu, M.D. Perez, P. Puyol, L. Sawyer, beta-lactoglobulin binds palmitate within its central cavity. J. Biol. Chem. 274(1), 170–174 (1999). https://doi.org/10.1074/jbc.274.1.170
A. Sivertsen, J. Isaksson, H.K. Leiros, J. Svenson, J.S. Svendsen, B.O. Brandsdal, Synthetic cationic antimicrobial peptides bind with their hydrophobic parts to drug site II of human serum albumin. BMC Struct. Biol.Struct. Biol. 14, 4 (2014). https://doi.org/10.1186/1472-6807-14-4
M. Muraki, K. Harata, N. Sugita, K. Sato, Origin of carbohydrate recognition specificity of human lysozyme revealed by affinity labeling. Biochemistry 35(42), 13562–13567 (1996). https://doi.org/10.1021/bi9613180
K. Harata, X-ray structure of monoclinic turkey egg lysozyme at 1.3 A resolution. Acta Crystallogr. D Biol. Crystallogr. Crystallogr. D Biol. Crystallogr. 49(Pt 5), 497–504 (1993). https://doi.org/10.1107/S0907444993005542
N. Noinaj, N.C. Easley, M. Oke, N. Mizuno, J. Gumbart, E. Boura, A.N. Steere, O. Zak, P. Aisen, E. Tajkhorshid, R.W. Evans, A.R. Gorringe, A.B. Mason, A.C. Steven, S.K. Buchanan, Structural basis for iron piracy by pathogenic Neisseria. Nature 483(7387), 53–58 (2012). https://doi.org/10.1038/nature10823
Y. Matsuda, A. Chakrabarti, K. Takahashi, K. Yamada, K. Nakata, T. Okuzumi, B.A. Mendelsohn, Chromatographic analysis of site-specific antibody-drug conjugates produced by AJICAP first-generation technology using a recombinant FcgammaIIIa receptor-ligand affinity column. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci.Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1177, 122753 (2021). https://doi.org/10.1016/j.jchromb.2021.122753
Y. Matsuda, T. Seki, K. Yamada, Y. Ooba, K. Takahashi, T. Fujii, S. Kawaguchi, T. Narita, A. Nakayama, Y. Kitahara, B.A. Mendelsohn, T. Okuzumi, Chemical site-specific conjugation platform to improve the pharmacokinetics and therapeutic index of antibody-drug conjugates. Mol. Pharm. 18(11), 4058–4066 (2021). https://doi.org/10.1021/acs.molpharmaceut.1c00473
T. Fujii, C. Reiling, C. Quinn, M. Kliman, B.A. Mendelsohn, Y. Matsuda, Physical characteristics comparison between maytansinoid-based and auristatin-based antibody-drug conjugates. Explor. Target Antitumor Ther. 2(6), 576–585 (2021). https://doi.org/10.37349/etat.2021.00064
H. Wu, D. Cao, T. Liu, J. Zhao, X. Hu, N. Li, Purification and characterization of recombinant human lysozyme from eggs of transgenic chickens. PLoS ONE 10(12), e0146032 (2015). https://doi.org/10.1371/journal.pone.0146032
J. Lescar, H. Souchon, P.M. Alzari, Crystal structures of pheasant and guinea fowl egg-white lysozymes. Protein Sci. 3(5), 788–798 (1994). https://doi.org/10.1002/pro.5560030508
D. Mercadante, L.D. Melton, G.E. Norris, T.S. Loo, M.A. Williams, R.C. Dobson, G.B. Jameson, Bovine beta-lactoglobulin is dimeric under imitative physiological conditions: dissociation equilibrium and rate constants over the pH range of 2.5–7.5. Biophys. J.. J. 103(2), 303–312 (2012). https://doi.org/10.1016/j.bpj.2012.05.041
M. Da Silva, S. Beauclercq, G. Harichaux, V. Labas, N. Guyot, J. Gautron, Y. Nys, S. Rehault-Godbert, The family secrets of avian egg-specific ovalbumin and its related proteins Y and X. Biol. Reprod.Reprod. 93(3), 71 (2015). https://doi.org/10.1095/biolreprod.115.130856
D. Fologea, B. Ledden, D.S. McNabb, J. Li, Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 91(5), 539011–539013 (2007). https://doi.org/10.1063/1.2767206
D.A. Belinskaia, P.A. Voronina, A.A. Batalova, N.V. Goncharov, Serum albumin. Encyclopedia 1(1), 65–75 (2020). https://doi.org/10.3390/encyclopedia1010009
F. Giansanti, L. Leboffe, F. Angelucci, G. Antonini, The nutraceutical properties of ovotransferrin and its potential utilization as a functional food. Nutrients 7(11), 9105–9115 (2015). https://doi.org/10.3390/nu7115453
M. Kallsten, R. Hartmann, K. Artemenko, S.B. Lind, F. Lehmann, J. Bergquist, Qualitative analysis of antibody-drug conjugates (ADCs): an experimental comparison of analytical techniques of cysteine-linked ADCs. Analyst 143(22), 5487–5496 (2018). https://doi.org/10.1039/c8an01178h
Z. Tawfiq, Y. Matsuda, M.J. Alfonso, C. Clancy, V. Robles, M. Leung, B.A. Mendelsohn, Analytical comparison of antibody-drug conjugates based on good manufacturing practice strategies. Anal. Sci. 36(7), 871–875 (2020). https://doi.org/10.2116/analsci.19P465
N. Shikida, S. Yamazaki, K. Takahashi, Y. Matsuda, K. Shimbo, Analytical studies on the conjugation site specificity of trastuzumab modified by Escherichia coli lipoate ligase A: multiple-enzyme digestion approach for peptide map**. Anal. Bioanal. Chem.Bioanal. Chem. 415, 6461–6469 (2023)
Y. Matsuda, Z. Tawfiq, M. Leung, B.A. Mendelsohn, Insight into temperature dependency and design of experiments towards process development for cysteine-based antibody-drug conjugates. ChemistrySelect 5(28), 8435–8439 (2020). https://doi.org/10.1002/slct.202001822
L. Conilh, L. Sadilkova, W. Viricel, C. Dumontet, Payload diversification: a key step in the development of antibody-drug conjugates. J. Hematol. Oncol.Hematol. Oncol. 16(1), 3 (2023). https://doi.org/10.1186/s13045-022-01397-y
T. Fujii, Y. Matsuda, S Novel formats of antibody conjugates: recent advances in payload diversity, conjugation, and linker chemistry. Expert Opin. Biol. Ther.Opin. Biol. Ther. 23(11), 1053–1065 (2023). https://doi.org/10.1021/jo2010186
I. Cheng-Sanchez, F. Moya-Utrera, C. Porras-Alcala, J.M. Lopez-Romero, F. Sarabia, Antibody-drug conjugates containing payloads from marine origin. Mar. Drugs 20(8), 494 (2022). https://doi.org/10.3390/md20080494
Acknowledgements
The authors would like to thank their colleagues from A**omoto Co., Inc. and A**omoto-Genetika Research Institute., as follows: Dr. Sergey Vasilievich Smirnov, Dr. Ilina Libovna Tokmakova, Mr. Kota Inoue, and Dr. Yasuhiro Mihara for LplA preparation; Dr. Uno Tagami for crystal structure modeling and constructive discussion; Ms. Natsuki Shikida and Dr. Kazutaka Shimbo for protein analysis; Dr. Shigeo Hirasawa, Dr. Mototaka Suzuki and Dr. Tatsuya Okuzumi for helpful comments and suggestions in this study.
Funding
This work was supported by A**omoto Co., Inc.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yamazaki, S., Takahashi, K. & Matsuda, Y. Tag-free protein modification by lipoate ligase A: exploring substrate tolerance. ANAL. SCI. 40, 1111–1119 (2024). https://doi.org/10.1007/s44211-024-00534-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s44211-024-00534-6