Abstract
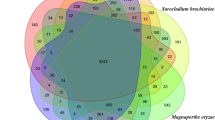
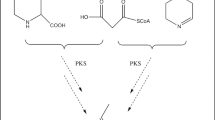
Sanghuangporus sanghuang is a medicinal macrofungus with antioxidant and antitumor activities, and it is enriched with secondary metabolites such as polysaccharides, terpenes, polyphenols, and styrylpyrone compounds. To explore the putative core genes and gene clusters involved in sanghuang biosynthesis, we sequenced and assembled a 40.5-Mb genome of S. sanghuang (SH1 strain). Using antiSMASH, local BLAST, and NCBI comparison, 12 terpene synthases (TPSs), 1 non-ribosomal peptide synthase, and five polyketide synthases (PKSs) were identified in SH1. Combining the transcriptome analysis with liquid chromatography mass spectrometry-ion trap-time of flight analysis, we determined that ShPKS1, one phenylalanine aminolyase (ShPAL), and one P450 monooxygenase (ShC4H1) were associated with hispidin biosynthesis. Structural domain comparison indicated that ShPKS2 and ShPKS3 are involved in the biosynthesis of orsellinic acid and 2-hydroxy-6-methylbenzoic acid, respectively. Furthermore, comparative genomic analysis of SH1 with 14 other fungi from the Hymenochaetaceae family showed variation in the number of TPSs among different genomes, with Coniferiporia weirii exhibiting only 9 TPSs and Inonotus obliquus having 20. The number of TPSs also differed among the genomes of three strains of S. sanghuang, namely Kangneng (16), MS2 (9), and SH1 (12). The type and number of PKSs also varied among species and even strains, ranging from two PKSs in Pyrrhoderma noxium to five PKSs in S. sanghuang SH1. Among the three strains of S. sanghuang, both the structural domains and the number of PKSs in strains MS2 and SH1 were consistent, whereas strain Kangneng exhibited only four PKSs and lacked the PKS with the structural domain KS-AT-DH-KR-ACP. Additionally, Sanghuangporus species exhibited more similar PKSs to Inonotus, with higher gene similarity around five PKSs, while showing differences from those of other fungi in the same family, including Phellinus lamaoensis. This result supports the independent taxonomic significance of the genus Sanghuangporus to some extent.









Similar content being viewed by others
Data Availability
This study project is available under NCBI BioProject accession number PRJNA960849 and BioSample accession number SAMN34331867. The complete genome assembly of Sanghuangporus sanghuang SH1 is available under GenBank accession number JASFXP000000000.
Abbreviations
- C. sulphurascens:
-
Coniferiporia sulphurascens
- C. weirii:
-
Coniferiporia weirii
- F. mediterranea:
-
Fomitiporia mediterranea
- G. junonius:
-
Gymnopilus junonius
- I. hispidus:
-
Inonotus hispidus
- I. obliquus:
-
Inonotus obliquus
- Pd. pouzarii:
-
Phellinidium pouzarii
- Pn. lamaoensis:
-
Phellinus lamaoensis
- Pl. nigrolimitatus:
-
Phellopilus nigrolimitatus
- Po. pini:
-
Porodaedalea pini
- Py. noxium:
-
Pyrrhoderma noxium
- S. baumii:
-
Sanghuangporus baumii
- S. sanghuang:
-
Sanghuangporus sanghuang
References
Joshi M et al (2021) Hymenochaetaceae. Mushrooms of Gujarat 3–11. https://doi.org/10.1007/978-981-16-4999-8_2
Tura D et al (2009) Medicinal species from genera Inonotus and Phellinus (Aphyllophoromycetideae): cultural-morphological peculiarities, growth characteristics, and qualitative enzymatic activity tests. Int J Med Mushrooms 11(3). https://doi.org/10.1615/intjmedmushr.v11.i3.100
Ikekawa T et al (1968) Antitumor action of some basidiomycetes, especially Phellinus linteus. GANN Japan J Cancer Res 59(2):155–157. https://doi.org/10.20772/cancersci1959.59.2_155
Wu S-H et al (2012) Species clarification for the medicinally valuable ‘sanghuang’ mushroom. Bot Stud 53(1)
Han J-G et al (2016) Species identity of Phellinus linteus (sanghuang) extensively used as a medicinal mushroom in Korea. J Microbiol 54:290–295. https://doi.org/10.1007/s12275-016-5520-2
Dai Y-C et al (2010) Current advances in Phellinus sensu lato: medicinal species, functions, metabolites and mechanisms. Appl Microbiol Biotechnol 87:1587–1593. https://doi.org/10.1007/s00253-010-2711-3
Li T et al (2023) Comparative compositions and activities of flavonoids from nine sanghuang strains based on solid-state fermentation and in vitro assays. Fermentation 9(3):308. https://doi.org/10.3390/fermentation9030308
Lin W-C et al (2017) Evaluation of antioxidant, anti-inflammatory and anti-proliferative activities of ethanol extracts from different varieties of Sanghuang species. RSC Adv 7(13):7780–7788. https://doi.org/10.1039/c6ra27198g
Hou R et al (2021) Chemical characterization of two fractions from Sanghuangporus sanghuang and evaluation of antidiabetic activity. J Funct Foods 87:104825. https://doi.org/10.1016/j.jff.2021.104825
Jiang W-P et al (2021) Sanghuangporus sanghuang mycelium prevents paracetamol-induced hepatotoxicity through regulating the MAPK/NF-κB, Keap1/Nrf2/HO-1, TLR4/PI3K/Akt, and CaMKKβ/LKB1/AMPK pathways and suppressing oxidative stress and inflammation. Antioxidants 10(6):897. https://doi.org/10.3390/antiox10060897
Liu K et al (2022) Phenolic composition and neuroprotective effects of the ethyl-acetate fraction from Inonotus sanghuang against H2O2-induced apoptotic cell death of primary cortical neuronal cells. Food Sci Biotechnol 31(9):1213–1223. https://doi.org/10.1039/c6ra27198g
Li IC et al (2020) Optimized production and safety evaluation of hispidin-enriched Sanghuangporus sanghuang mycelia. Food Sci Nutr 8(4):1864–1873. https://doi.org/10.1002/fsn3.1469
Lee I-K, Yun B-S (2011) Styrylpyrone-class compounds from medicinal fungi Phellinus and Inonotus spp., and their medicinal importance. J Antibiot 64(5):349–359. https://doi.org/10.1038/ja.2011.2
Palkina KA et al (2021) Therapeutic potential of hispidin—fungal and plant polyketide. J Fungi 7(5):323. https://doi.org/10.3390/jof7050323
Gonindard C et al (1997) Synthetic hispidin, a PKC inhibitor, is more cytotoxic toward cancer cells than normal cells in vitro. Cell Biol Toxicol 13:141–153. https://doi.org/10.1023/A:1007321227010
Lee I-K et al (2008) Chemical constituents of Gymnopilus spectabilis and their antioxidant activity. Mycobiology 36(1):55–59. https://doi.org/10.4489/MYCO.2008.36.1.055
Shao Y et al (2020) The genome of the medicinal macrofungus Sanghuang provides insights into the synthesis of diverse secondary metabolites. Front Microbiol 10:3035. https://doi.org/10.3389/fmicb.2019.03035/full
Wangun HVK et al (2006) Inotilone and related phenylpropanoid polyketides from Inonotus sp. and their identification as potent COX and XO inhibitors. Org Biomol Chem 4(13):2545–2548. https://doi.org/10.1039/b604505g
Li I et al (2022) Pilot study: Nutritional and preclinical safety investigation of fermented hispidin-enriched sanghuangporus sanghuang mycelia: a promising functional food material to improve sleep. Front Nutr 8:788965. https://doi.org/10.3389/fnut.2021.788965/full
Tu PTB, Tawata S (2014) Anti-obesity effects of hispidin and Alpinia zerumbet bioactives in 3T3-L1 adipocytes. Molecules 19(10):16656–16671. https://doi.org/10.3390/molecules191016656
Park I-H et al (2004) A β-secretase (BACE1) inhibitor hispidin from the mycelial cultures of Phellinus linteus. Planta Med 70(02):143–146. https://doi.org/10.1055/s-2004-815491
Zhang H et al (2019) The integration of metabolome and proteome reveals bioactive polyphenols and hispidin in ARTP mutagenized Phellinus baumii. Sci Rep 9(1):16172. https://doi.org/10.1038/s41598-019-52711-7
Perrin P, Towers G (1973) Hispidin biosynthesis in cultures of Polyporus hispidus. Phytochemistry 12(3):589–592. https://doi.org/10.1016/s0031-9422(00)84448-9
Wang Z-X et al (2022) Diverse metabolites and pharmacological effects from the basidiomycetes Inonotus hispidus. Antibiotics 11(8):1097. https://doi.org/10.3390/antibiotics11081097
Schubert M, Lindgreen S, Orlando L (2016) AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes 9(1). https://doi.org/10.1186/s13104-016-1900-2
Bankevich A et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Walker BJ et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. https://doi.org/10.1371/journal.pone.0112963
Seppey M, Manni M, Zdobnov EM (2019) BUSCO: assessing genome assembly and annotation completeness, in Gene prediction. Springer New York: New York, NY. p. 227-245
Haas BJ et al (2008) Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol 9:1–22. https://doi.org/10.1186/gb-2008-9-1-r7
Chen C et al (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49(W1):W293–W296. https://doi.org/10.1093/nar/gkab301
Herbst DA, Townsend CA, Maier T (2018) The architectures of iterative type I PKS and FAS. Nat Prod Rep 35(10):1046–1069. https://doi.org/10.1039/c8np00039e
Shen N et al (2022) Plant flavonoids: classification, distribution, biosynthesis, and antioxidant activity. Food Chem 383:132531. https://doi.org/10.1016/j.foodchem.2022.132531
Pluskal T et al (2019) The biosynthetic origin of psychoactive kavalactones in kava. Nat Plants 5(8):867–878. https://doi.org/10.1038/s41477-019-0474-0
Nabavi SM et al (2020) Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnol Adv 38:107316. https://doi.org/10.1016/j.biotechadv.2018.11.005
Panche AN, Diwan AD, Chandra SR (2016) Flavonoids: an overview. J Nutr Sci 5:e47. https://doi.org/10.1017/jns.2016.41
Atanasoff-Kardjalieff AK et al (2023) Biosynthesis of the isocoumarin derivatives fusamarins is mediated by the PKS8 gene cluster in fusarium. ChemBioChem 24(6):e202200342. https://doi.org/10.1002/cbic.202200342
Wang S et al (2022) Mushrooms do produce flavonoids: metabolite profiling and transcriptome analysis of flavonoid synthesis in the medicinal mushroom Sanghuangporus baumii. J Fungi 8(6):582. https://doi.org/10.3390/jof8060582
Zhang H et al (2022) A fungal NRPS-PKS enzyme catalyses the formation of the flavonoid naringenin. Nat Commun 13(1):6361. https://doi.org/10.1038/s41467-022-34150-7
Zhang W et al (2023) Discovery of a unique flavonoid biosynthesis mechanism in fungi by genome mining. Angew Chem 135(12):e202215529. https://doi.org/10.1002/ange.202215529
Palkina KA et al (2023) Domain truncation in hispidin synthase orthologs from non-bioluminescent fungi does not lead to hispidin biosynthesis. Int J Mol Sci 24(2):1317. https://doi.org/10.3390/ijms24021317
Chepkirui C et al (2018) An unprecedented spiro [furan-2, 1’-indene]-3-one derivative and other nematicidal and antimicrobial metabolites from Sanghuangporus sp. (Hymenochaetaceae, Basidiomycota) collected in Kenya. Phytochem Lett 25:141–146. https://doi.org/10.1016/j.phytol.2018.04.022
Fiasson J-L, Niemelä T (1984) The Hymenochaetales: a revision of the European poroid taxa. Karstenia 24(1):14–28. https://doi.org/10.29203/ka.1984.224
Kaur N et al (2019) Investigations on antioxidative potential of poroid medicinal mushroom Porodaedalea pini (Agaricomycetes). Int J Med Mushrooms 21(6). https://doi.org/10.1615/intjmedmushrooms.2019030883
Lee I-K, Yun B-S (2007) Highly oxygenated and unsaturated metabolites providing a diversity of hispidin class antioxidants in the medicinal mushrooms Inonotus and Phellinus. Bioorg Med Chem 15(10):3309–3314. https://doi.org/10.1016/j.bmc.2007.03.039
Sułkowska-Ziaja K, Balik M, Muszyńska B (2021) Selected species of the genus phellinus–chemical composition, biological activity, and medicinal applications. Chem Biodivers 18(11):e2100609. https://doi.org/10.1002/cbdv.202100609
Zan LF et al (2015) A new antioxidant pyrano [4, 3-c][2] benzopyran-1, 6-dione derivative from the medicinal mushroom Fomitiporia ellipsoidea. Nat Prod Commun 10(2):1934578X1501000226. https://doi.org/10.1177/1934578X1501000226
Jiang J-H, Wu S-H, Zhou L-W (2021) The first whole genome sequencing of sanghuangporus sanghuang provides insights into its medicinal application and evolution. J Fungi. https://doi.org/10.3390/jof7100787
Shen Z-Q et al (2023) Genome re-annotation and transcriptome analyses of Sanghuangporus sanghuang. J Fungi. https://doi.org/10.3390/jof9050505
Mitiouchkina T et al (2020) Plants with genetically encoded autoluminescence. Nat Biotechnol 38(8):944–946. https://doi.org/10.1038/s41587-020-0500-9
Zhou L-W et al (2016) Global diversity and taxonomy of the Inonotus linteus complex (Hymenochaetales, Basidiomycota): Sanghuangporus gen. nov., Tropicoporus excentrodendri and T. guanacastensis gen. et spp. nov., and 17 new combinations. Fungal Diver 77:335–347. https://doi.org/10.1007/s13225-015-0335-8
Lu Y et al (2022) Genomic comparative analysis of cordyceps pseudotenuipes with other species from cordyceps. Metabolites 12(9):844. https://doi.org/10.3390/metabo12090844
Shen S et al (2021) Addressing widespread misidentifications of traditional medicinal mushrooms in Sanghuangporus (Basidiomycota) through ITS barcoding and designation of reference sequences. IMA Fungus 12(1). https://doi.org/10.1186/s43008-021-00059-x
Zhou Q et al (2021) Deep sequencing of the Sanghuangporus vaninii transcriptome reveals dynamic landscapes of candidate genes involved in the biosynthesis of active compounds. Arch Microbiol 203(5):2315–2324. https://doi.org/10.1007/s00203-021-02225-6
Zhao Z et al (2013) Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 14(1). https://doi.org/10.1186/1471-2164-14-274
Richardson SM et al (2022) BioWF: a naturally-fused, di-domain biocatalyst from biotin biosynthesis displays an unexpectedly broad substrate scope. ChemBioChem 23(17):e202200171. https://doi.org/10.1002/cbic.202200171
Awakawa T et al (2013) Pyranonigrin E: a PKS-NRPS hybrid metabolite from Aspergillus niger identified by genome mining. ChemBioChem 14(16):2095–2099. https://doi.org/10.1002/cbic.201300430
Theobald S, Vesth TC, Andersen MR (2019) Genus level analysis of PKS-NRPS and NRPS-PKS hybrids reveals their origin in Aspergilli. BMC Genomics 20(1):1–12. https://doi.org/10.1186/s12864-019-6114-2
Wang Y et al (2021) Bioactive PKS–NRPS alkaloids from the plant-derived endophytic fungus Xylaria arbuscula. Molecules 27(1):136. https://doi.org/10.3390/molecules27010136
Quin MB et al (2017) Spatial organization of multi-enzyme biocatalytic cascades. Org Biomol Chem 15(20):4260–4271. https://doi.org/10.1039/c7ob00391a
Nielsen ML et al (2016) Linker flexibility facilitates module exchange in fungal hybrid PKS-NRPS engineering. PLoS One 11(8):e0161199. https://doi.org/10.1371/journal.pone.0161199
Acknowledgements
We acknowledge TopEdit LLC for the linguistic editing and proofreading during the preparation of this manuscript.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (31860177), General Project of Basic Research Program in Yunnan Province (202101AT070218), the Reserve Talents for Young and Middle-aged Academic and Technical Leaders of the Yunnan Province (202205AC160044).
Author information
Authors and Affiliations
Contributions
Conceptualization, JZ and YW; methodology, YW, XYW, and ZWL; software, YW, XLY, and DW; formal analysis, ZWL; investigation, JMN and XLY; resources, YY; writing—original draft preparation, XYW; writing—review and editing, XYW, YW, and JZ.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
**nyue Wang and Jiansheng Wei have contributed equally to this work.
Responsible Editor: Celia Maria de Almeida Soares
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Wei, J., Liu, Z. et al. Comparative genomic analysis of Sanghuangporus sanghuang with other Hymenochaetaceae species. Braz J Microbiol 55, 87–100 (2024). https://doi.org/10.1007/s42770-023-01212-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-01212-x