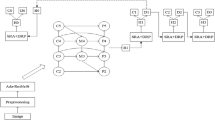
Abstract
Osteoporotic Vertebral Fracture (OVFs) is a common lumbar spine disorder that severely affects the health of patients. With a clear bone blocks boundary, CT images have gained obvious advantages in OVFs diagnosis. Compared with CT images, X-rays are faster and more inexpensive but often leads to misdiagnosis and miss-diagnosis because of the overlap** shadows. Considering how to transfer CT imaging advantages to achieve OVFs classification in X-rays is meaningful. For this purpose, we propose a multi-modal semantic consistency network which could do well X-ray OVFs classification by transferring CT semantic consistency features. Different from existing methods, we introduce a feature-level mix-up module to get the domain soft labels which helps the network reduce the domain offsets between CT and X-ray. In the meanwhile, the network uses a self-rotation pretext task on both CT and X-ray domains to enhance learning the high-level semantic invariant features. We employ five evaluation metrics to compare the proposed method with the state-of-the-art methods. The final results show that our method improves the best value of AUC from 86.32 to 92.16%. The results indicate that multi-modal semantic consistency method could use CT imaging features to improve osteoporotic vertebral fracture classification in X-rays effectively.










Similar content being viewed by others
Data Availability
The data that support the findings of this study are not openly available due to human data and are available from the corresponding author upon reasonable request.
References
Carberry, G. A., Pooler, B. D., Binkley, N., Lauder, T. B., Bruce, R. J., & Pickhardt, P. J. (2013). Unreported vertebral body compression fractures at abdominal multidetector CT. Radiology, 268, 120–126.
Ren, S., Guo, K. H., Ma, J. Q., Zhu, F. H., Hu, B., & Zhou, H. M. (2021). Realistic medical image super-resolution with pyramidal feature multi-distillation networks for intelligent healthcare systems. Neural Computing and Applications, 1–16.
Sevli, O. (2021). A deep convolutional neural network-based pigmented skin lesion classification application and experts evaluation. Neural Computing and Applications, 33, 12039–12050.
Shakeel, P. M., Burhanuddin, M. A., & Desa, M. I. (2020). Automatic lung cancer detection from CT image using improved deep neural network and ensemble classifier. Neural Computing and Applications, 1–14.
Bar, A., Wolf, L., Amitai, O. B., Toledano, E., Elnekave, E. (2017). Compression fractures detection on CT. In Medical imaging 2017 computer-aided diagnosis. USA: Orlando, Florida.
Tomita, N., Cheung, Y. Y., & Hassanpour, S. (2018). Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Computers in Biology and Medicine, 98, 8–15.
Fang, Y. J., Li, W., Chen, X. J., Chen, K. M., Kang, H., Yu, P. X., & Li, S. L. (2021). Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. European Radiology, 31, 1831–1842.
Cha, K. H., Hadjiiski, L., Samala, R. K., Chan, H. P., Caoili, E. M., & Cohan, R. H. (2016). Urinary bladder segmentation in CT urography using deep-learning convolutional neural network and level sets. Medical Physics, 43, 1882–1896.
Frighetto-Pereira, L., Menezes-Reis, R., Metzner, G. A., Rangayyan, R. M., Azevedo-Marques, P. M., & Nogueira-Barbosa, M. H. (2015). Semiautomatic classification of benign versus malignant vertebral compression fractures using texture and gray-level features in magnetic resonance images. In 2015 IEEE 28th International Symposium on Computer-Based Medical Systems (pp. 88–92). Sao Carlos, Brazil.
Burns, J. E., Yao, J., & Summers, R. M. (2017). Vertebral body compression fractures and bone density: Automated detection and classification on CT images. Radiology, 284, 788–797.
Bar, A., Wolf, L., Amitai, O. B., Toledano, E., & Elnekave, E. (2017). Compression fractures detection on CT. In Medical imaging 2017: Computer-aided diagnosis(Vol. 10134, p. 1013440). Orlando, Florida, USA.
Tomita, N., Cheung, Y. Y., & Hassanpour, S. (2018). Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Computers in Biology and Medicine, 98, 8–15.
Zhang, W. C., Ouyang, W. L., Li, W., & Xu, D. (2018). Collaborative and adversarial network for unsupervised domain adaptation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 3801–3809), Salt Lake City, UT, USA.
Tran, L., Sohn, K., Yu, X., Liu, X., & Chandraker, M. K. (2018). Joint pixel and feature-level domain adaptation in the wild. ar**v preprint ar**v:1803.00068.
Ghifary, M., Kleijn, W. B., Zhang, M., Balduzzi, D., & Li, W. (2016). Deep reconstruction-classification networks for unsupervised domain adaptation. In European conference on computer vision (pp. 597–613), Amsterdam, The Netherlands.
Tzeng, E., Hoffman, J., Saenko, K., & Darrell, T. (2017). Adversarial discriminative domain adaptation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 7167–7176), Honolulu, HI, USA.
Tsai, Y. H., Hung, W. C., Schulter, S., Sohn, K., Yang, M. H., & Chandraker, M. (2018). Learning to adapt structured output space for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 7472–7481), Salt Lake City, UT, USA.
Tzeng, E., Hoffman, J., Zhang, N., Saenko, K., & Darrell, T. (2014). Deep domain confusion: Maximizing for domain invariance. ar**v preprint ar**v:1412.3474.
Sun, B., & Saenko, K. (2016). Deep coral: Correlation alignment for deep domain adaptation. In European conference on computer vision (pp. 443–450). Amsterdam, The Netherlands.
Long, M. S., Cao, Y., Wang, J. M., & Jordan, M. (2015). Learning transferable features with deep adaptation networks. In International conference on machine learning (pp. 97–105). Lille, France.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., & Ozair, S., et al. (2014). Generative adversarial nets. Advances in Neural Information Processing Systems, 27.
JHoffman, J., Tzeng, E., Park, T., Zhu, J. Y., Isola, P., & Saenko, K., et al. (2018). Cycada: Cycle-consistent adversarial domain adaptation. In International conference on machine learning (pp. 1989–1998). Stockholm, Sweden.
Chen, Y. H., Chen, W. Y., Chen, Y. T., Tsai, B. C., Frank Wang, Y. C., & Sun, M. (2017). No more discrimination: Cross city adaptation of road scene segmenters. In Proceedings of the IEEE international conference on computer vision (pp. 1992–2001). Venice, Italy.
Sankaranarayanan, S., Balaji, Y., Jain, A., Lim, S. N., & Chellappa, R. (2017). Unsupervised domain adaptation for semantic segmentation with gans. ar**v preprint ar**v:1711.06969.
Zhang, Y. H., Qiu, Z. F., Yao, T., Liu, D., & Mei, T. (2018). Fully convolutional adaptation networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 6810–6818), Salt Lake City, UT, USA.
Dou, Q., Ouyang, C., Chen, C., Chen, H., & Heng, P. A. (2018). Unsupervised cross-modality domain adaptation of convnets for biomedical image segmentations with adversarial loss. ar**v preprint ar**v:1804.10916.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., & Fei-Fei, L. (2015). Imagenet large scale visual recognition challenge. International Journal of Computer Vision, 115, 211–252.
Mahajan, D., Girshick, R., Ramanathan, V., He, K., Paluri, M., & Li, Y., et al. (2018). Exploring the limits of weakly supervised pretraining. In Proceedings of the European conference on computer vision (pp. 181–196), Munich, Germany.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234–241), Munich, Germany.
Li, W., Zhao, Y. F., Chen, X., **ao, Y., & Qin, Y. Y. (2018). Detecting Alzheimer’s disease on small dataset: A knowledge transfer perspective. IEEE Journal of Biomedical and Health Informatics, 23, 1234–1242.
Laun, F. B., Lederer, W., Daniel, H., Kuder, T. A., Delorme, S., & Schlemmer, H. P., et al. (2018). Domain adaptation for deviating acquisition protocols in cnn-based lesion classification on diffusion-weighted MR images. In Image analysis for moving organ, breast, and thoracic images: Third international workshop, Granada, Spain.
Javanmardi, M., & Tasdizen, T. (2018). Domain adaptation for biomedical image segmentation using adversarial training. In 2018 IEEE 15th international symposium on biomedical imaging (pp. 554–558), Washington, DC, USA.
Dou, Q., Ouyang, C., Chen, C., Chen, H., Glocker, B., Zhuang, X. H., & Heng, P. A. (2019). Pnp-adanet: Plug-and-play adversarial domain adaptation network at unpaired cross-modality cardiac segmentation. IEEE Access, 7, 99065–99076.
Ganin, Y., Ustinova, E., Ajakan, H., Germain, P., Larochelle, H., Laviolette, F., & Lempitsky, V. (2016). Domain-adversarial training of neural networks. The Journal of Machine Learning Research, 17, 2030–2096.
Zhu, Y. C., Zhuang, F. Z., Wang, J. D., Chen, J. W., Shi, Z. P., Wu, W. J., & He, Q. (2019). Multi-representation adaptation network for cross-domain image classification. Neural Networks, 119, 214–221.
Zhu, Y. C., Zhuang, F. Z., Wang, J. D., Ke, G. L., Chen, J. W., Bian, J., & He, Q. (2020). Deep subdomain adaptation network for image classification. IEEE Transactions on Neural Networks and Learning Systems, 32, 1713–1722.
Xu, M. H., Zhang, J., Ni, B. B., Li, T., Wang, C. J., Tian, Q., & Zhang, W. J. (2020). Adversarial domain adaptation with domain mixup. In Proceedings of the AAAI conference on artificial intelligence (pp. 6502–6509), New York City, NY, USA.
Ganin, Y., & Lempitsky, V. (2015). Unsupervised domain adaptation by backpropagation. In International conference on machine learning (pp. 1180–1189), Lille, France.
Löffler, M. T., Sekuboyina, A., Jacob, A., Grau, A. L., Scharr, A., El Husseini, M., et al. (2020). A vertebral segmentation dataset with fracture grading. Radiology: Artificial Intelligence, 2, e190138.
Sekuboyina, A., Husseini, M. E., Bayat, A., Löffler, M., Liebl, H., Li, H., & Kirschke, J. S. (2021). VerSe: A Vertebrae labelling and segmentation benchmark for multi-detector CT images. Medical Image Analysis, 73, 102166.
Kingma, D. P., & Ba, J. (2014). Adam: A method for stochastic optimization. ar**v preprint ar**v:1412.6980.
He, K. M., Zhang, X. Y., Ren, S. Q., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770–778), Las Vegas, NV, USA.
Donahue, J., Jia, Y., Vinyals, O., Hoffman, J., Zhang, N., Tzeng, E., & Darrell, T. (2014). Decaf: A deep convolutional activation feature for generic visual recognition. In International conference on machine learning (pp. 647–655), Bei**g, China.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (U21A20390), the National Key Research and Development Program of China (2018YFC2001302), the Development Project of Jilin Province of China (nos. 20200801033GH, 20200403172SF, YDZJ202101ZYTS128), Jilin Provincial Key Laboratory of Big Data Intelligent Computing (no. 20180622002JC), The Fundamental Research Funds for the Central University, JLU.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest. This research study was conducted retrospectively from data obtained for clinical purposes. We consulted extensively with the Clinical Research Ethics Committee of the Second Hospital of Jilin University who determined that our study did not need ethical approval.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Y., Bai, T., Li, T. et al. Osteoporotic Vertebral Fracture Classification in X-rays Based on a Multi-modal Semantic Consistency Network. J Bionic Eng 19, 1816–1829 (2022). https://doi.org/10.1007/s42235-022-00234-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42235-022-00234-9