Abstract
Objectives
The process of measuring morphological parameters of the cervical spine through computed tomography (CT) is repetitive and time-consuming. Deep learning (DL) improves efficiency and consistency. In this study, we built a series of DL-based segmentation algorithms to automatically measure five key parameters for the evaluation of cervical spondylotic myelopathy. Subsequently, we compared the performance of our algorithm with that of physicians to assess its accuracy and clinical application value.
Methods
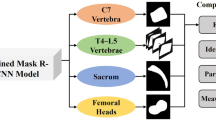
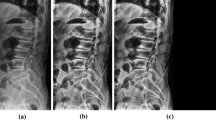
Cervical spine CT images of 685 patients were divided into a training (n = 548) and a test set (n = 137). The training set was used to develop a VB-Net DL model, including a 3D segmentation model of multiple cervical spine subregions and a key point location model on the midsagittal slice of the cervical spine CT. The parameters measured included sagittal vertebral canal diameter (SCD), sagittal vertebral body diameter (VBD), Pavlov’s ratio, transverse vertebral canal diameter (TCD), and osseous spinal canal area (OSCA). Manual measurements were performed by a radiologist and a spinal surgeon. The model’s performance was evaluated using the Mann–Whitney U test, Pearson correlation, mean absolute error, and Bland–Altman plots.
Results
DL and manual measurements of the Pavlov’s ratio, SCD, VBD, TCD, and OSCA in the test set showed similar accuracy and consistency. The VB-Net model’s Pearson correlation coefficient exceeded 0.8 for most parameters.
Conclusions
Our VB-Net-based DL approach can effectively approximate the manual measurements of cervical CT morphological parameters in humans, offing physicians an accurate and efficient auxiliary diagnostic tool.





Similar content being viewed by others
Data availability
The data sets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DL:
-
Deep learning
- CSS:
-
Cervical spinal stenosis
- CSM:
-
Cervical spondylotic myelopathy
- DCS:
-
Developmental cervical stenosis
- SCD:
-
Sagittal canal diameter
- TCD:
-
Transverse canal diameter
- VBD:
-
Vertebral body diameter
- OSCA:
-
Osseous spinal canal area
- OPLL:
-
Ossification of the posterior longitudinal ligament
References
Ailon T, Smith JS, Shaffrey CI, et al. Outcomes of operative treatment for adult cervical deformity: a prospective multicenter assessment with 1-year follow-up. Neurosurgery. 2018;83:1031–9. https://doi.org/10.1093/neuros/nyx574.
Toki S, Higashino K, Manabe H, et al (2021) Morphometric analysis of subaxial cervical spine with myelopathy: a comparison with the normal population. Spine Surg Relat Res 5:34–40.https://doi.org/10.22603/ssrr.2020-0061
Pavlov H, Torg JS, Robie B, Jahre C. Cervical spinal stenosis: determination with vertebral body ratio method. Radiology. 1987;164:771–5. https://doi.org/10.1148/radiology.164.3.3615879.
Aebli N, Wicki AG, Rüegg TB, et al. The Torg-Pavlov ratio for the prediction of acute spinal cord injury after a minor trauma to the cervical spine. Spine J. 2013;13:605–12. https://doi.org/10.1016/j.spinee.2012.10.039.
Kumar A, Sahu S, Sethi S, et al. Computerized tomography-based morphometric analysis of cervical spinal canal in central indian population. J Neurosci Rural Pract. 2020;11:274–7. https://doi.org/10.1055/s-0040-1703962.
Song K-J, Choi B-W, Kim S-J, et al. The relationship between spinal stenosis and neurological outcome in traumatic cervical spine injury: an analysis using Pavlov’s ratio, spinal cord area, and spinal canal area. Clin Orthop Surg. 2009;1:11–8. https://doi.org/10.4055/cios.2009.1.1.11.
Chen X, Wang X, Zhang K, et al. Recent advances and clinical applications of deep learning in medical image analysis. Med Image Anal. 2022;79: 102444. https://doi.org/10.1016/j.media.2022.102444.
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. https://doi.org/10.1038/nature14539.
Zheng Q, Shellikeri S, Huang H, et al. Deep learning measurement of leg length discrepancy in children based on radiographs. Radiology. 2020;296:152–8. https://doi.org/10.1148/radiol.2020192003.
Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39:1137–49. https://doi.org/10.1109/TPAMI.2016.2577031.
von Schacky CE, Wilhelm NJ, Schäfer VS, et al. Multitask deep learning for segmentation and classification of primary bone tumors on radiographs. Radiology. 2021;301:398–406. https://doi.org/10.1148/radiol.2021204531.
Choi H, Kim YK, Yoon EJ, et al. Cognitive signature of brain FDG PET based on deep learning: domain transfer from Alzheimer’s disease to Parkinson’s disease. Eur J Nucl Med Mol Imaging. 2020;47:403–12. https://doi.org/10.1007/s00259-019-04538-7.
Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:640–51. https://doi.org/10.1109/TPAMI.2016.2572683.
Tajbakhsh N, Jeyaseelan L, Li Q, et al. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med Image Anal. 2020;63: 101693. https://doi.org/10.1016/j.media.2020.101693.
Balakrishnan G, Zhao A, Sabuncu MR, et al. VoxelMorph: a learning framework for deformable medical image registration. IEEE Trans Med Imaging. 2019. https://doi.org/10.1109/TMI.2019.2897538.
Zou J, Gao B, Song Y, Qin J. A review of deep learning-based deformable medical image registration. Front Oncol. 2022;12:1047215. https://doi.org/10.3389/fonc.2022.1047215.
Zhao S, Wu X, Chen B, Li S. Automatic spondylolisthesis grading from MRIs across modalities using faster adversarial recognition network. Med Image Anal. 2019;58: 101533. https://doi.org/10.1016/j.media.2019.101533.
Pang S, Su Z, Leung S, et al. Direct automated quantitative measurement of spine by cascade amplifier regression network with manifold regularization. Med Image Anal. 2019;55:103–15. https://doi.org/10.1016/j.media.2019.04.012.
Alukaev D, Kiselev S, Mustafaev T, et al. A deep learning framework for vertebral morphometry and Cobb angle measurement with external validation. Eur Spine J. 2022;31:2115–24. https://doi.org/10.1007/s00586-022-07245-4.
Nai R, Liu X, et al. Automatic measurement of the patellofemoral joint parameters in the Laurin view: a deep learning-based approach. Eur Radiol. 2023;33:566–77. https://doi.org/10.1007/s00330-022-08967-1.
Zhang S-C, Sun J, Liu C-B, et al. Clinical application of artificial intelligence-assisted diagnosis using anteroposterior pelvic radiographs in children with developmental dysplasia of the hip. Bone Joint J. 2020;102B:1574–81. https://doi.org/10.1302/0301-620X.102B11.BJJ-2020-0712.R2.
Kim Y-T, Jeong TS, Kim YJ, et al. Automatic spine segmentation and parameter measurement for radiological analysis of whole-spine lateral radiographs using deep learning and computer vision. J Digit Imaging. 2023. https://doi.org/10.1007/s10278-023-00830-z.
Lee HM, Kim YJ, Cho JB, et al. Computer-aided diagnosis for determining sagittal spinal curvatures using deep learning and radiography. J Digit Imaging. 2022;35:846–59. https://doi.org/10.1007/s10278-022-00592-0.
Yoo HJ, Kim YJ, Hong H, et al. Deep learning-based fully automated body composition analysis of thigh CT: comparison with DXA measurement. Eur Radiol. 2022;32:7601–11. https://doi.org/10.1007/s00330-022-08770-y.
Funding
This study received funding from Bei**g Natural Science Foundation (Z190020), National Natural Science Foundation of China (81971578, 81901791, 82102638), Peking University Third Hospital Clinical Key Project (BYSYZD2021040, BYSY2018003, BYSYZD2019005), Peking University Third Hospital Clinical Cohort Construction Project (BYSYDL2022007), Peking University Health Science Center Education and Teaching Research Project (2021YB05), Peking University Teaching and Learning 2.0 (2023YB01).
Author information
Authors and Affiliations
Contributions
All authors contributed extensively to the work presented in this paper. Their roles are given below. Conception and design: HQO, YL. Administrative support: NL, HSY. Provision of study materials or patients: HQO, ST, YL. Collection and assembly of data: ELZ, HCP, YTH, DLH, SYD. Data analysis and interpretation: XML,PD. Manuscript writing: all authors. Final approval of manuscript: all authors.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Institutional review board statement
The methods were performed in accordance with relevant guidelines and regulations and approved by Peking University Third Hospital Medical Science Research Ethic Committee.
Informed consent statement
Patient consent was waived due to retrospective study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, Y., Zhang, E., Ouyang, H. et al. Application of deep learning in analysing morphological parameters of cervical computed tomography scans. Chin J Acad Radiol 7, 50–57 (2024). https://doi.org/10.1007/s42058-024-00136-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42058-024-00136-1