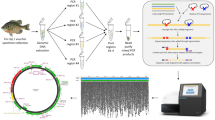
Abstract
Environmental DNA (eDNA) is a promising biomonitoring tool for marine ecosystems, but its effectiveness for North Pacific coastal fishes is limited by the inability of existing barcoding primers to differentiate among rockfishes in the genus Sebastes. Comprised of 110 commercially and ecologically important species, this recent radiation is exceptionally speciose, and exhibits high sequence similarity among species at standard barcoding loci. Here, we report new Sebastes-specific metabarcoding primers that target mitochondrial cytochrome B. Amongst the 110 Sebastes species, 85 unique barcodes (of which 62 are species-specific) were identified in our amplicon region based on available reference sequences. The majority of the remaining barcodes are shared by only two species. Importantly, MiSebastes yield unique barcodes for 28 of 44 commercially harvested species in California, a dramatic improvement compared to the widely employed MiFish-U 12 S primers which only recover one of 44. Tests of these primers in an aquarium mesocosm containing 16 rockfish species confirms the utility of these new primers for eDNA metabarcoding, providing an important biomonitoring tool for these key coastal marine fishes.


Similar content being viewed by others
Data Availability
Sanger sequence Cytochrome B barcode data generated for this study are publicly available on NCBI GenBank, bioproject number PRJNA733285. The MiFish-U and MiSebastes sequencing datasets are available on Dryad (https://doi.org/10.5068/D1J951).
Code availability
Code used for primer design and subsequent analyses are publicly available here: https://github.com/markusmin/MiSebastes.
References
Altschul SF, Gish W, Miller W et al (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Andruszkiewicz EA, Koseff JR, Fringer OB et al (2019) Modeling environmental DNA transport in the coastal ocean using Lagrangian particle tracking. Front Mar Sci 6:1–14. https://doi.org/10.3389/fmars.2019.00477
Bohmann K, Evans A, Gilbert MTP et al (2014) Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 29:358–367. https://doi.org/10.1016/j.tree.2014.04.003
Boyer F, Mercier C, Bonin A et al (2016) obitools: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16:176–182. https://doi.org/10.1111/1755-0998.12428
Brown EA, Chain FJJ, Crease TJ et al (2015) Divergence thresholds and divergent biodiversity estimates: Can metabarcoding reliably describe zooplankton communities? Ecol Evol 5:2234–2251. https://doi.org/10.1002/ece3.1485
Bru D, Martin-Laurent F, Philippot L (2008) Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl Environ Microbiol 74:1660–1663. https://doi.org/10.1128/AEM.02403-07
Bucklin A, Lindeque PK, Rodriguez-Ezpeleta N et al (2016) Metabarcoding of marine zooplankton: Prospects, progress and pitfalls. J Plankton Res 38:393–400. https://doi.org/10.1093/plankt/fbw023
Closek CJ, Santora JA, Starks HA et al (2019) Marine Vertebrate Biodiversity and Distribution Within the Central California Current Using Environmental DNA (eDNA) Metabarcoding and Ecosystem Surveys. Front Mar Sci 6:1–17. https://doi.org/10.3389/fmars.2019.00732
Crooks G, Hon G, Chandonia J, Brenner S (2004) NCBI GenBank FTP Site\nWebLogo: a sequence logo generator. Genome Res 14:1188–1190. https://doi.org/10.1101/gr.849004.1
Curd EE, Gold Z, Kandlikar GS et al (2019) Anacapa Toolkit: an environmental DNA toolkit for processing multilocus metabarcode datasets. Methods Ecol Evol 10:1469–1475
Deiner K, Bik HM, Mächler E et al (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol Ecol 26:5872–5895. https://doi.org/10.1111/mec.14350
Dick EJ, MacCall AD (2014) Status and productivity of cowcod, Sebastes levis, in the Southern California Bight, 2013. Natl Ocean Atmos Adm Natl Mar Fish Serv
Doi H, Fukaya K, Oka S, ichiro et al (2019) Evaluation of detection probabilities at the water-filtering and initial PCR steps in environmental DNA metabarcoding using a multispecies site occupancy model. Sci Rep 9:1–8. https://doi.org/10.1038/s41598-019-40233-1
Doney SC, Ruckelshaus M, Emmett Duffy J et al (2012) Climate change impacts on marine ecosystems. Ann Rev Mar Sci 4:11–37
Faircloth BC, Glenn TC (2014) Protocol: Preparation of an AMPure XP substitute (AKA Serapure). DOI 10:J9MW2F26
Ficetola GF, Miaud C, Pompanon F, Taberlet P (2008) Species detection using environmental DNA from water samples. Biol Lett 4:423–425. https://doi.org/10.1098/rsbl.2008.0118
Ficetola GF, Coissac E, Zundel S et al (2010) Taberlet P Pompanon F An In silico approach for the evaluation of DNA barcodes. BMC Genom. https://doi.org/10.1186/1471-2164-11-434
Gao X, Lin H, Revanna K, Dong Q (2017) A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 18:247
Gillett DJ, Pondella DJ, Freiwald J et al (2012) Comparing volunteer and professionally collected monitoring data from the rocky subtidal reefs of southern California, USA. Environ Monit Assess 184:3239–3257. https://doi.org/10.1007/s10661-011-2185-5
Gold ZJ (2020) Design and implementation of environmental DNA metabarcoding methods for monitoring the Southern. California marine protected area network
Gold Z, Curd E, Goodwin K et al (2021) Improving Metabarcoding Taxonomic Assignment: A Case Study of Fishes in a Large Marine Ecosystem. Authorea Prepr. https://doi.org/10.22541/au.161407483.33882798/v1
Hyde JR, Vetter RD (2007) The origin, evolution, and diversification of rockfishes of the genus Sebastes (Cuvier). Mol Phylogenet Evol 44:790–811. https://doi.org/10.1016/j.ympev.2006.12.026
Ingram T, Kai Y (2014) The Geography of Morphological Convergence in the Radiations of Pacific Sebastes Rockfishes. Am Nat 184:E115–E131. https://doi.org/10.1086/678053
Ji Y, Ashton L, Pedley SM et al (2013) Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol Lett 16:1245–1257. https://doi.org/10.1111/ele.12162
Jo T, Murakami H, Masuda R et al (2017) Rapid degradation of longer DNA fragments enables the improved estimation of distribution and biomass using environmental DNA. Mol Ecol Resour 17:e25–e33. https://doi.org/10.1111/1755-0998.12685
Kearse M, Moir R, Wilson A et al (2012) Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kelly RP, Port JA, Yamahara KM et al (2014a) Harnessing DNA to improve environmental management. Science 344:1455–1456. https://doi.org/10.1126/science.1251156
Kelly RP, Port JA, Yamahara KM, Crowder LB (2014) Using environmental DNA to census marine fishes in a large mesocosm. PLoS One. https://doi.org/10.1371/journal.pone.0086175
Klindworth A, Pruesse E, Schweer T et al (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. https://doi.org/10.1093/nar/gks808
Lea RN, McAllister RD, VenTresca DA (1999) Biological aspects of nearshore rockfishes of the genus Sebastes from central California: with notes on ecologically related sport fishes. State of California, The Resources Agency, Department of Fish and Game
Love MS, Yoklavich M, Thorsteinson LK (2002) The rockfishes of the northeast Pacific. Univ of California Press
Mata VA, Rebelo H, Amorim F et al (2019) How much is enough? Effects of technical and biological replication on metabarcoding dietary analysis. Mol Ecol 28:165–175. https://doi.org/10.1111/mec.14779
Meyer RS, Curd EE, Schweizer T et al (2019) The California environmental DNA “CALeDNA” program. bioRxiv 503383
Mills KL, Laidig T, Ralston S, Sydeman WJ (2007) Diets of top predators indicate pelagic juvenile rockfish (Sebastes spp.) abundance in the California Current System. Fish Oceanogr 16:273–283
Miya M, Sato Y, Fukunaga T et al (2015) MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes : detection of more than 230 subtropical marine species Subject Category. Subject Areas
Miya M, Minamoto T, Yamanaka H et al (2016) Use of a Filter Cartridge for Filtration of Water Samples and Extraction of Environmental DNA. J Vis Exp. https://doi.org/10.3791/54741
Murphy HM, Jenkins GP (2010) Observational methods used in marine spatial monitoring of fishes and associated habitats: a review. Mar Freshw Res 61:236. https://doi.org/10.1071/mf09068
O’Donnell JL, Kelly RP, Shelton AO et al (2017) Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ 5:e3044. https://doi.org/10.7717/peerj.3044
Port JA, O’Donnell JL, Romero-Maraccini OC et al (2016) Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol Ecol 25:527–541. https://doi.org/10.1111/mec.13481
Riaz T, Shehzad W, Viari A et al (2011) EcoPrimers: Inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res 39:1–11. https://doi.org/10.1093/nar/gkr732
Schliep KP (2011) phangorn: Phylogenetic analysis in R. Bioinformatics 27:592–593. https://doi.org/10.1093/bioinformatics/btq706
Spens J, Evans AR, Halfmaerten D et al (2017) Comparison of capture and storage methods for aqueous macrobial eDNA using an optimized extraction protocol: advantage of enclosed filter. Methods Ecol Evol 8:635–645
Stat M, Huggett MJ, Bernasconi R et al (2017) Ecosystem biomonitoring with eDNA: Metabarcoding across the tree of life in a tropical marine environment. Sci Rep 7:1–11. https://doi.org/10.1038/s41598-017-12501-5
Taberlet P, Coissac E, Hajibabaei M, Rieseberg L (2012) Environmental DNA. Mol Ecol 21:1789–1793
Team RC (2013) R: A language and environment for statistical computing
Thompson AR, Hyde JR, Watson W et al (2016) Rockfish assemblage structure and spawning locations in southern California identified through larval sampling. Mar Ecol Prog Ser 547:177–192. https://doi.org/10.3354/meps11633
Thomsen PF, Willerslev E (2015) Environmental DNA - An emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18. https://doi.org/10.1016/j.biocon.2014.11.019
Thomsen PF, Møller PR, Sigsgaard EE et al (2016) Environmental DNA from Seawater Samples Correlate with Trawl Catches of Subarctic, Deepwater Fishes. PLoS One 11:e0165252. https://doi.org/10.1371/journal.pone.0165252
Thorson JT, Wetzel C (2015) The status of canary rockfish (Sebastes pinniger) in the California Current in 2015. Natl Ocean Atmos Adm Natl Mar Fish Serv
Truelove NK, Andruszkiewicz EA, Block BA (2019) A rapid environmental DNA method for detecting white sharks in the open ocean. Methods Ecol Evol 10:1128–1135. https://doi.org/10.1111/2041-210X.13201
Untergasser A, Nijveen H, Rao X et al (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35:71–74. https://doi.org/10.1093/nar/gkm306
Ushio M, Murata K, Sado T et al (2018) Demonstration of the potential of environmental DNA as a tool for the detection of avian species. Sci Rep 8:1–10. https://doi.org/10.1038/s41598-018-22817-5
Valentini A, Taberlet P, Miaud C et al (2016) Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol Ecol 25:929–942. https://doi.org/10.1111/mec.13428
Valsecchi E, Bylemans J, Goodman SJ et al (2020) Novel universal primers for metabarcoding environmental DNA surveys of marine mammals and other marine vertebrates. Environ DNA. https://doi.org/10.1002/edn3.72
Wetzel CR, Cronin-Fine L, Johnson KF (2017) Status of Pacific ocean perch (Sebastes alutus) along the US west coast in 2017. Pacific Fish Manag Counc 7700:97220
Wildlife CD of F and (2020) Final California Commercial Landings for 2019
Williams GD, Levin PS, Palsson WA (2010) Rockfish in Puget Sound: An ecological history of exploitation. Mar Policy 34:1010–1020
Yamamoto S, Masuda R, Sato Y et al (2017) Environmental DNA metabarcoding reveals local fish communities in a species-rich coastal sea. Sci Rep 7:1–12. https://doi.org/10.1038/srep40368
Acknowledgements
We thank Emily Curd and Meixi Lin for their assistance with the primer design process. We thank Andrew Thompson and Dovi Kacev for providing us with DNA extracted from tissue samples. We thank Adam Wall for his help processing our Sanger sequencing data. We thank Keira Monuki, McKenzie Koch, Beverly Shih, Cristopher Ruano, and Nikita Sridhar for their assistance with sample preparation in the lab.
Funding
Grants to MAM from the UCLA Undergraduate Research Scholars Program, to ZJG from the US-NSF Graduate Research Fellowship (DEG No. 1650604), and to PHB from the Howard Hughes Medical Institute Science Education Program supported this study.
Author information
Authors and Affiliations
Contributions
Contributions include: Conceptualization MAM, PHB, ZJG. Investigation MAM, ZJG. Funding Acquisition MAM, PHB, ZJG. Data Curation MAM, ZJG. Formal Analysis MAM, ZJG. Writing – Original Draft Preparation MAM, PHB, ZJG.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Consent to participate
No consent to participate was required for this study.
Consent for publication
All authors consent to the publication of this manuscript.
Ethical approval
No ethics approval was needed for this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
12686_2021_1219_MOESM1_ESM.eps
Electronic supplementary material 1 Fig. 1 Detections of the 11 species identified by the MiSebastes primers by individual technical replicates from the kelp forest aquarium at the California Science Center. Samples are divided into “bottom” and “surface” samples. Biological replicates are indicated by the numbers in the sample names; technical replicates are indicated by the lettering following the biological replicate numbers (EPS 18 kb)
12686_2021_1219_MOESM2_ESM.xlsx
Electronic supplementary material 2 Table 1 DNA sequences for Sanger sequencing of tissue samples barcoded with the MiSebastes primer set, along with the following BLAST results: top three hits for each sequence, percent identity, query cover, and accession number of the matching sequences (XLSX 16 kb)
12686_2021_1219_MOESM3_ESM.xlsx
Electronic supplementary material 3 Supplementary Table 2 Amplicons for the region targeted by the MiSebastes primer set as predicted by in silico PCR results from ecoPCR. For each amplicon, the species that share the amplicon, the region where the species are found, and the number of mismatches in the priming regions are listed (XLSX 17 kb)
12686_2021_1219_MOESM4_ESM.xlsx
Electronic supplementary material 4 Supplementary Table 3 Inventory of the species in the kelp forest aquarium at the California Science Center, along with detections by MiFish-U (using both the Fish CARD and CRUX databases) and the MiSebastes primer sets (XLSX 17 kb)
12686_2021_1219_MOESM5_ESM.xlsx
Electronic supplementary material 5 Supplementary Table 4 List of all samples in each of the two sequencing libraries in this study (one each for MiSebastes and MiFish-U), along with associated metadata (XLSX 15 kb)
Rights and permissions
About this article
Cite this article
Min, M.A., Barber, P.H. & Gold, Z. MiSebastes: An eDNA metabarcoding primer set for rockfishes (genus Sebastes). Conservation Genet Resour 13, 447–456 (2021). https://doi.org/10.1007/s12686-021-01219-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-021-01219-2