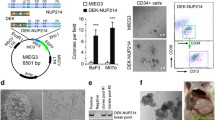
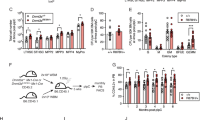
Abstract
Cancer is a very rare event at the cellular level, although it is a common disease at the body level as one third of humans die of cancer. A small subset of cells in the body harbor the cellular features that constitute a permissive window for a particular genetic change to induce cancer. The significance of a permissive window is ironically best shown by a large number of failures in generating the animal model for acute myeloid leukemia (AML) with t(8;21). Over the decades, the RUNX1-ETO fusion gene created by t(8;21) has been introduced into various types of hematopoietic cells, largely at adult stage, in mice; however, all the previous attempts failed to generate tractable AML models. In stark contrast, we recently succeeded in inducing AML with the clinical features seen in human patients by specifically introducing RUNX1-ETO in childhood hematopoietic stem cells (HSCs). This result in mice is consistent with adolescent and young adult (AYA) onset in human t(8;21) patients, and suggests that childhood HSCs constitute the permissive window for RUNX1-ETO leukemogenesis. If loss of a permissive window is induced pharmacologically, cancer cells might be selectively targeted. Such a permissive window modifier may serve as a novel therapeutic drug.


Similar content being viewed by others
Data availability
The data supporting the findings of this review are available within the article and its references, or from the corresponding author upon reasonable request.
References
Watt SM, Hua P, Roberts I. Increasing complexity of molecular landscapes in human hematopoietic stem and progenitor cells during development and aging. Int J Mol Sci. 2022;23(7):3675.
Mancarella D, Plass C. Epigenetic signatures in cancer: proper controls, current challenges and the potential for clinical translation. Genome Med. 2021;13(1):23.
Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497.
Rowley JD. A new consistent chromosomal abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine fluorescence and Giemsa Staining. Nature. 1973;243(5405):290–3.
Groffen J, Stephenson J, Heisterkamp N, Deklein A, Bartram C, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93–9.
Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. NEJM. 2001;344(14):1038–42.
Meier R, Greve G, Zimmer D, Bresser H, Berberich B, Langova R, et al. The antileukemic activity of decitabine upon PML/RARA-negative AML blasts is supported by all-trans retinoic acid: in vitro and in vivo evidence for cooperation. Blood Cancer J. 2022;12(8):1–3.
Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88(23):10431–4.
Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, et al. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology. 1993;194(1):314–31.
Wang SW, Speck NA. Purification of core-binding factor, a protein that binds the conserved core site in murine leukemia virus enhancers. Mol Cell Biol. 1992;12(1):89–102.
Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, et al. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15(3):303–6.
Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23(24):4284–96.
Lang L. Barry Marshall 2005 Nobel Laureate in Medicine and Physiology: Anil K. Rustgi, MD, Section Editor. Gastroenterology. 2005;129(6):1813–4.
Yi Z, Yuan Z. From discovery to cure, a great journey of the Hepatitis C Virus study. J Infect Dis Immun. 2022;2(02):109–12.
Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, et al. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–9.
Chin DW, Watanabe-Okochi N, Wang CQ, Tergaonkar V, Osato M. Mouse models for core binding factor leukemia. Leukemia. 2015;29(10):1970–80.
Cabezas Wallscheid N, Eichwald V, de Graaf J, Löwer M, Lehr HA, Kreft A, et al. Instruction of haematopoietic lineage choices, evolution of transcriptional landscapes and cancer stem cell hierarchies derived from an AML 1ETO mouse model. EMBO Mol Med. 2013;5(12):1804–20.
Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ, et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA. 2001;98(18):10398–403.
Shields CL, Bas Z, Laiton A, Silva AM, Sheikh A, Lally SE, et al. Retinoblastoma: emerging concepts in genetics, global disease burden, chemotherapy outcomes, and psychological impact. Eye. 2022;25:1–8.
Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30(13):1492–502.
Gandarillas A. The mysterious human epidermal cell cycle, or an oncogene-induced differentiation checkpoint. Cell Cycle. 2012;11(24):4507–16.
Sieber OM, Tomlinson SR, Tomlinson IPM. Tissue, cell and stage specificity of (epi)mutations in cancers. Nat Rev Cancer. 2005;5(8):649–55.
Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018;37(1):173–87.
Feldser DM, Kostova KK, Winslow MM, Taylor SE, Cashman C, Whittaker CA. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–5.
Levine AJ, Jenkins NA, Copeland NG. The roles of initiating truncal mutations in human cancers: the order of mutations and tumor cell type matters. Cancer Cell. 2019;35(1):10–5.
López-Otín C, Pietrocola F, Roiz-Valle D, Galluzzi L, Kroemer G. Meta-hallmarks of aging and cancer. Cell Metab. 2023;35(1):12–35.
Qiu B, Matthay KK. Advancing therapy for neuroblastoma. Nat Rev Clin Oncol. 2022;25:1–9.
Roberts I, Fordham NJ, Rao A, Bain BJ. Neonatal leukaemia. Br J Haematol. 2018;182(2):170–84.
Yanagida M, Osato M, Yamashita N, Liqun H, Jacob B, Wu F, et al. Increased dosage of Runx1/AML1 acts as a positive modulator of myeloid leukemogenesis in BXH2 mice. Oncogene. 2005;24(28):4477–85.
Osato M, Ito Y. Increased dosage of the RUNX1/AML1 gene: a third mode of RUNX leukemia? Crit Rev Eukaryot Gene Expr. 2005;15(3):217–28.
Hamaguchi Y, Kondoh T, Fukuda M, Yamasaki K, Yoshiura KI, Moriuchi H, et al. Leukopenia, macrocytosis, and thrombocytopenia occur in young adults with Down syndrome. Gene. 2022;835:146663.
Labuhn M, Perkins K, Varghese L, Garnett C, Papaemmanuil E, Metzner M, et al. Mechanisms of progression of myeloid preleukemia to transformed myeloid leukemia in children with Down syndrome. Cancer Cell. 2019;12;36(2):123–38.
Garnett C, Cruz Hernandez D, Vyas P. GATA1 and cooperating mutations in myeloid leukaemia of Down syndrome. IUBMB Life. 2020;72(1):119–30.
Bolouri H, Farrar JE, Triche T Jr, Ries RE, Lim EL, Alonzo TA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat Med. 2018;24(1):103–12.
Abdallah MG, Niibori-Nambu A, Morii M, Yokomizo T, Yokomizo T, Ideue T, et al. RUNX1-ETO (RUNX1-RUNX1T1) induces myeloid leukemia in mice in an age-dependent manner. Leukemia. 2021;35(10):2983–8.
Downing JR. The core-binding factor leukemias: lessons learned from murine models. Curr Opin Genet Dev. 2003;13(1):48–54.
Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y, et al. A previously unidentified alternatively spliced isoform of t (8; 21) transcript promotes leukemogenesis. Nat Med. 2006;12(8):945–9.
Traver D, Akashi K, Weissman IL, Lagasse E. Mice defective in two apoptosis pathways in the myeloid lineage develop acute myeloblastic leukemia. Immunity. 1998;9(1):47–57.
Koh CP, Bahirvani AG, Wang CQ, Yokomizo T, Ng CE, Du L, Tergaonkar V, et al. Highly efficient Runx1 enhancer eR1-mediated genetic engineering for fetal, child and adult hematopoietic stem cells. Gene. 2022;851:147049.
Ng CE, Yokomizo T, Yamashita N, Cirovic B, ** H, Wen Z, et al. A Runx1 intronic enhancer marks hemogenic endothelial cells and hematopoietic stem cells. Stem cells. 2010;28(10):1869–81.
Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116(10):2808–16.
Kumar A, D’Souza SS, Thakur AS. Understanding the journey of human hematopoietic stem cell development. Stem Cells Int. 2019;2019:2141475.
Wang CQ, Mok MM, Yokomizo T, Tergaonkar V, Osato M. Runx family genes in tissue stem cell dynamics. RUNX Proteins in Development and Cancer. Adv Exp Med Biol. 2017;962:117–38.
Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470–83.
Bai J, Yokomizo-Nakano T, Kubota S, Sun Y, Kanai A, Iimori M. Overexpression of Hmga2 activates Igf2bp2 and remodels transcriptional program of Tet2-deficient stem cells in myeloid transformation. Oncogene. 2021;40(8):1531–41.
Yokomizo T, Ideue T, Morino-Koga S, Tham CY, Sato T, Takeda N. Independent origins of fetal liver haematopoietic stem and progenitor cells. Nature. 2022;609(7928):779–84.
Hock H, Meade E, Medeiros S, Schindler JW, Valk PJ, Fujiwara Y, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18(19):2336–41.
Hock H, Shimamura A. ETV6 in hematopoiesis and leukemia predisposition. Semin Hematol. 2017;54(2):98–104.
Van der Meer LT, Jansen JH, Van Der Reijden BA. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 2010;24(11):1834–43.
Tsang JC, Yu Y, Burke S, Buettner F, Wang C, Kolodziejczyk AA, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol. 2015;16(1):1–6.
Kruta M, Sunshine MJ, Chua BA, Fu Y, Chawla A, Dillingham CH, et al. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell Stem Cell. 2021;28(11):1950–65.
Ye M, Zhang H, Amabile G, Yang H, Staber PB, Zhang P, et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol. 2013;15(4):385–94.
Ikeda K, Ueda T, Yamasaki N, Nakata Y, Sera Y, Nagamachi A, et al. Maintenance of the functional integrity of mouse hematopoiesis by EED and promotion of leukemogenesis by EED haploinsufficiency. Sci Rep. 2016;6(1):1–3.
**e H, Xu J, Hsu JH, Nguyen M, Fujiwara Y, Peng C, et al. Polycomb repressive complex 2 regulates hematopoietic stem cell maintenance and differentiation in a developmental stage-specific manner. Cell Stem Cell. 2014;14(1):68.
Tanaka-Yano M, Wang D, Meader E, Kinney MA, Morris V, da Rocha EL, et al. Lin28b-Let-7-PRC1 axis guides developmental maturation of the hematopoietic system. Blood. 2021;138:21.
Jones M, Chase J, Brinkmeier M, Xu J, Weinberg DN, Schira J. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J Clin Invest. 2015;125(5):2007–20.
Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–5.
Cesana M, Guo MH, Cacchiarelli D, Wahlster L, Barragan J, Doulatov S, et al. A CLK3-HMGA2 alternative splicing axis impacts human hematopoietic stem cell molecular identity throughout development. Cell Stem Cell. 2018;22(4):575-588.e7.
Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–6.
Hilpert M, Legrand C, Bluteau D, Balayn N, Betems A, Bluteau O, et al. p19INK4d controls hematopoietic stem cells in a cell-autonomous manner during genotoxic stress and through the microenvironment during aging. Stem Cell Rep. 2014;3(6):1085–102.
Magee JA, Ikenoue T, Nakada D, Lee JY, Guan KL, Morrison SJ. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11(3):415–28.
Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512(7513):198–202.
Li Y, Kong W, Yang W, Patel RM, Casey EB, Okeyo-Owuor T. Single-cell analysis of neonatal HSC ontogeny reveals gradual and uncoordinated transcriptional reprogramming that begins before birth. Cell Stem Cell. 2020;27(5):732–47.
Florian MC, Nattamai KJ, Dörr K, Marka G, Überle B, Vas V. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503(7476):392–6.
Oshima M, Hasegawa N, Mochizuki-Kashio M, Muto T, Miyagi S, Koide S, et al. Ezh2 regulates the Lin28/let-7 pathway to restrict activation of fetal gene signature in adult hematopoietic stem cells. Exp Hematol. 2016;44(4):282–96.
Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, et al. The Lin28b–let-7–Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15(8):916–25.
Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140(4):445–9.
Du QY, Zhu ZM, Pei DS. The biological function of IGF2BPs and their role in tumorigenesis. Invest New Drugs. 2021;39(6):1682–93.
Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–8.
Wang S, Chim B, Su Y, Khil P, Wong M, Wang X, et al. Enhancement of LIN28B-induced hematopoietic reprogramming by IGF2BP3. Genes Dev. 2019;33(15–16):1048–68.
Lederer M, Bley N, Schleifer C, Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. 2014;29:3–12.
Xue L, Cai JY, Ma J, Huang Z, Guo MX, Fu LZ. Global expression profiling reveals genetic programs underlying the developmental divergence between mouse and human embryogenesis. BMC Genom. 2013;14:1–7.
Acknowledgements
This work was supported by National Medical Research Council (MOH-CIRG21nov-0007), Joint NCIS and NUS Centre for Cancer Research (N2CR) Seed Funding Programme, the National Research Foundation Singapore, the Singapore Ministry of Education under its Research Centres of Excellence initiative, and JSPS Kakenhi Grants (JP15H04312, JP16K14613, 18KT0026, 19K07668, and 22H02904).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Abdallah, M.G., Teoh, V.S.I., Dutta, B. et al. Childhood hematopoietic stem cells constitute the permissive window for RUNX1-ETO leukemogenesis. Int J Hematol 117, 830–838 (2023). https://doi.org/10.1007/s12185-023-03605-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12185-023-03605-y