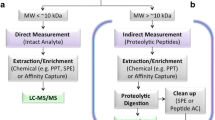
Abstract
Genetically modified organisms (GMOs) express one or more exogenous genes inserted through genetic engineering resulting in the production of novel protein(s). Agencies responsible for deregulating GMOs require that levels of the newly expressed protein(s) be characterized. Ideally, the quantitation methods used will be validated according to Good Laboratory Practices (GLPs) that are accepted by these global regulatory agencies. Targeted protein quantitation may be performed using enzyme-linked immunosorbent assays (ELISAs) with commercial kits or specially produced and validated antibodies. Proteins may also be quantitated by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Although some scientific guidance exists for validation of ELISA, the need remains for a harmonized set of protocols for protein quantitation in testing of GMOs, particularly for LC-MS/MS. We surveyed various industry experts to determine current practices for assay linearity, accuracy, and precision; assay specificity; detection limits; and methods to assess analyte stability for both ELISA and LC-MS/MS. We compared the survey results with assay criteria suggested in the literature and governmental guidance documents.
Similar content being viewed by others
References
AOAC (1998) Peer-verified methods program. Manual on policies and procedures. http://citeseerx.ist.psu.edu/viewdoc/download;jsessionid=C3D82F9AE00CA520AF2E3F46E67E8D6E?doi=10.1.1.196.7223&rep=rep1&type=pdf. Accessed 25 Mar 2015
Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B (2007) Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem 389(4):1017–1031
Bonfini L, Angers-Loustau A, Petrillo M, Ciabatti I, Gatto F, Rosa S, Lievans A, Kreysa J (2016) The European Union reference methods database and decision supporting tool for the analysis of genetically modified organisms: GMOMETHODS and JRC GMO-matrix. In: Watson RR, Preedy VR (eds) Genetically modified organisms in food. Elsevier, London, pp. 275–288
CAC/GL 74 (2010) http://www.codexalimentarius.org/standards/list-of-standards. Accessed 25 Mar 2015
CropLife International (2011a) http://www.detection-methods.com/. Accessed 15 Dec 2015
CropLife International (2011b) Detection methods basics http://www.detection-methods.com/detection-methods-basics. Accessed 22 June 2015
Czyz M, Dembczynski R, Marecik R, Wojas-Turek J, Milczarek M, Pajtasz-Piasecka E, Wiertrzyk J, and Pniewski T (2014) Freeze-drying of plant tissue containing HBV surface antigen for the oral vaccine against hepatitis B. Biomed Res Int. doi:10.1155/2014/485689
Doran PM (2006) Foreign protein degradation and instability in plants and plant tissue cultures. Trends in Biotechnol 24(9):426–432
ENGL Working Group on “Method Verification” (2011) Verification of analytical methods for GMO testing when implementing interlaboratory validated methods. Publications of the European Union, Luxembourg
EPA (1996) Residue chemistry test guidelines. OPPTS 860.1340 residue analytical method
European Commission (2010) Guidance document on pesticide residue analytical methods. SANCO/825/00 rev. 8.1 16/11/2010
European Medicines Agency (2011) Guideline on bioanalytical method validation. http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2011/08/WC500109686.pdf. Accessed 25 Mar 2015
FDA (2013) Guidance for industry: bioanalytical method validation. 2013 http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm368107.pdf. Accessed 25 Mar 2015
Fernandez A, Mills ENC, Lovik M, Spoek A, Germini A, Mikalsen A, Wal JM (2013) Endogenous allergens and compositional analysis in the allergenicity assessment of genetically modified plants. Food Chem Toxicol 62:1–6
Findlay JWA, Dillard RF (2007) Appropriate calibration curve fitting in ligand binding assays. AAPS J 9(2):E260–E267
Gong CY, Wang T (2013) Proteomic evaluation of genetically modified crops: current status and challenges. Front Plant Science 4:1–8
Gosling JP (1990) A decade of development in immunoassay methodology. Clin Chem 36(8):1408–1427
Grothaus GD, Bandla M, Currier T, Giroux R, Jenkins GR, Lipp M, Shan G, Stave JW, Pantellam V (2006) Immunoassay as an analytical tool in agricultural biotechnology. Journal AOAC International 89(4):913–928
Hale JE (2013) Advantageous Uses of Mass Spectrometry for the Quantification of Proteins. Int J Proteomics 2013:1-6
Herman RA, Shan G (2011) Data interpretation and sources of error. In: Shan G (ed) Immunoassays in agricultural biotechnology. Wiley, Hoboken, pp. 189–218
Hill RC, Oman TJ, Shan G, Schafer B, Eble J, Chen C (2015) Development and validation of a multiplexed protein quantitation assay for the determination of the three recombinant proteins in soybean tissues by liquid chromatography with tandem mass spectrometry. J Ag Food Chem 63:7450–7461
Hu XT, Owens MA (2011) Multiplexed protein quantification in maize leaves by liquid chromatography coupled to tandem mass spectrometry: an alternative tool to immunoassays for target protein analysis in genetically engineered crops. J Ag Food Chem 59:3551–3558
International Standard ISO 21572 (2013) Food stuffs—molecular biomarker analysis—protein based methods. Second ed
Kamle S, Ojha A, Kumar A (2011) Development of an enzyme linked immunosorbant assay for the detection of Cry2Ab protein in transgenic plants. GM Crops 2(2):118–125
Lipp M, Anklam E, Stave JW (2000) Validation of an immunoassay for detection and quantitation of a genetically modified soybean in food and food fractions using reference materials: interlaboratory study. Journal AOAC International 83(4):919–927
Lipton CR, Dautlick JX, Grothaus GD, Hunst PL, Magin KM, Mihaliak CA, Rubio FM, Stave JW (2000) Guidelines for the validation and use of immunoassays for determination of introduced proteins in biotechnology enhanced crops and derived food ingredients. Food Agr Immunol 12(2):153–164
Ocaña MF, Fraser PD, Patel RK, Halket JM, Bramley PM (2007) Mass spectrometric detection of CP4 EPSPS in genetically modified soya and maize. Rapid Commun Mass Spectrom 2(3):319–328
Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2(1):43–50
Privalle LS, Chen J, Clapper G, Hunst P, Spiegelhalter F, Zhong CX (2012) Development of an agricultural biotechnology crop product: testing from discovery to commercialization. J Agr Food Chem 60(41):10179–10187
SANCO Guidelines 2009 Method validation and quality control procedures for pesticide residues analysis in food and feed. Document NO. SANCO/10684/2009
Schauzu M (2013) The European Union’s regulatory framework on genetically modified organisms and derived foods and feeds. Adv Gen Eng 2:109. doi:10.4172/2169-0111.1000109
Schmidt J, Alarcon C (2011) Immunoassay method development. In: Shan G (ed) Immunoassays in agricultural biotechnology. Wiley, Hoboken, pp. 115–138
Shillito R, Currier T (2011) Immunoassays as a GM detection method in international trade. In: Shan G (ed) Immunoassays in agricultural biotechnology. Wiley, Hoboken, pp. 309–324
US Department of Health and Human Services, Food and Drug Administration (2013) Guidance for industry bioanalytical method validation revision 1. http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM368107.pdf. Accessed 25 Mar 2015
Acknowledgments
The authors thank the Analytical Excellence through Industry Collaboration (AEIC) organization for providing a venue for discussion and a helpful review of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
No outside funding was received for this study.
Conflict of Interest
Sharon B. Settlage declares she has no conflicts of interest. Julie E. Eble declares she has no conflicts of interest. Jayant K. Bhanushali declares he has no conflicts of interest. Matthew L. Cheever declares he has no conflicts of interest. Ai-Guo Gao declares he has no conflicts of interest. David A. Goldstrohm declares he has no conflicts of interest. Ryan Hill declares he has no conflicts of interest. Tiger X. Hu declares he has no conflicts of interest. Charles R. Powley declares he has no conflicts of interest. Anita Unger declares she has no conflicts of interest. Guomin Shan declares he has no conflicts of interest.
Ethical Approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed Consent
Not applicable.
Rights and permissions
About this article
Cite this article
Settlage, S.B., Eble, J.E., Bhanushali, J.K. et al. Validation Parameters for Quantitating Specific Proteins Using ELISA or LC-MS/MS: Survey Results. Food Anal. Methods 10, 1339–1348 (2017). https://doi.org/10.1007/s12161-016-0689-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12161-016-0689-x