Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease characterized by dysfunction of the upper and lower motor neurons resulting in muscle weakness and wasting. Recently, several studies on ALS patients and ALS animal models indicated that intramuscular toxicity played a role in ALS disease progression; however, the mechanisms driving this are unknown. In this study, we explored the possible dysfunction of lipid metabolism in myocytes associated with ALS. Initially, skeletal muscle from 41 ALS patients, as well as 53 non-ALS control subjects, was investigated, and we identified that lipid droplet accumulation in the muscle fibers of ALS patients was significantly increased, especially in patients with FUS mutations. A myoblast (C2C12) cell line expressing mutant FUS (FUS-K510Q) was able to induce lipid droplet accumulation and mitochondrial dysfunction. Consistently, transgenic flies expressing FUS-K510Q under a muscle-specific driver showed elevated triglyceride levels in the flight muscles, as well as locomotor defects. Biochemical analysis of C2C12 cells and fly muscle tissues showed upregulation of PLIN2, and downregulation of ATGL and CPT1A, indicating inhibition of lipolysis and fatty acid β-oxidation in muscle cells with FUS mutations. Our study provided a potential explanation for the pathogenesis associated with lipid droplets accumulating in skeletal muscle in ALS. Our data also suggested that disordered lipid metabolism and mitochondrial dysfunction play a crucial role in intramuscular toxicity in ALS.






Similar content being viewed by others
Data Availability
The datasets analyzed in this study are available from the corresponding author on reasonable request.
References
Statland JM, Barohn RJ, McVey AL, Katz JS, Dimachkie MM (2015) Patterns of weakness, classification of motor neuron disease, and clinical diagnosis of sporadic amyotrophic lateral sclerosis. Neurol Clin 33(4):735–748. https://doi.org/10.1016/j.ncl.2015.07.006
Chia R, Chiò A, Traynor BJ (2018) Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 17(1). https://doi.org/10.1016/S1474-4422(17)30401-5
Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC (2011) Amyotrophic lateral sclerosis. Lancet 377(9769):942–955. https://doi.org/10.1016/S0140-6736(10)61156-7
Menon P, Kiernan MC, Vucic S (2014) ALS pathophysiology: insights from the split-hand phenomenon. Clin Neurophysiol 125(1):186–193. https://doi.org/10.1016/j.clinph.2013.07.022
Dupuis L, Echaniz-Laguna A (2010) Skeletal muscle in motor neuron diseases: therapeutic target and delivery route for potential treatments. Curr Drug Targets 11(10):1250–1261. https://doi.org/10.2174/1389450111007011250
Picchiarelli G, Demestre M, Zuko A, Been M, Higelin J, Dieterlé S, Goy M-A, Mallik M et al (2019) FUS-mediated regulation of acetylcholine receptor transcription at neuromuscular junctions is compromised in amyotrophic lateral sclerosis. Nat Neurosci 22(11):1793–1805. https://doi.org/10.1038/s41593-019-0498-9
Zhou B, Wang H, Cai Y, Wen H, Wang L, Zhu M, Chen Y, Yu Y et al (2020) FUS P525L mutation causing amyotrophic lateral sclerosis and movement disorders. Brain Behav 10(6):e01625. https://doi.org/10.1002/brb3.1625
Farese RV, Walther TC (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139(5):855–860. https://doi.org/10.1016/j.cell.2009.11.005
Angelini C, Nascimbeni AC, Cenacchi G, Tasca E (2016) Lipolysis and lipophagy in lipid storage myopathies. Biochim Biophys Acta 1862(7):1367–1373. https://doi.org/10.1016/j.bbadis.2016.04.008
Chaves-Filho AB, Pinto IFD, Dantas LS, Xavier AM, Inague A, Faria RL, Medeiros MHG, Glezer I et al (2019) Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis. Sci Rep 9(1):11642. https://doi.org/10.1038/s41598-019-48059-7
Scaricamazza S, Salvatori I, Giacovazzo G, Loeffler JP, Renè F, Rosina M, Quessada C, Proietti D et al (2020) Skeletal-muscle metabolic reprogramming in ALS-SOD1 mice predates disease onset and is a promising therapeutic target. iScience 23(5):101087. https://doi.org/10.1016/j.isci.2020.101087
Deng J, Wang P, Chen X, Cheng H, Liu J, Fushimi K, Zhu L, Wu JY (2018) FUS interacts with ATP synthase beta subunit and induces mitochondrial unfolded protein response in cellular and animal models. Proc Natl Acad Sci U S A 115(41):E9678–E9686. https://doi.org/10.1073/pnas.1806655115
Bollinger LM, Campbell MS, Brault JJ (2018) Palmitate and oleate co-treatment increases myocellular protein content via impaired protein degradation. Nutrition 46:41–43. https://doi.org/10.1016/j.nut.2017.07.017
Capel F, Cheraiti N, Acquaviva C, Hénique C, Bertrand-Michel J, Vianey-Saban C, Prip-Buus C, Morio B (2016) Oleate dose-dependently regulates palmitate metabolism and insulin signaling in C2C12 myotubes. Biochim Biophys Acta 1861(12 Pt A):2000–2010. https://doi.org/10.1016/j.bbalip.2016.10.002
Liu X, Chen J, Li X, Gao S, Deng M (2014) Generation of induced pluripotent stem cells from amyotrophic lateral sclerosis patientcarrying SOD1-V14M mutation. Zhonghua Yi Xue Za Zhi 94(27):2143–2147
Deng J, Wu W, **e Z, Gang Q, Yu M, Liu J, Wang Q, Lv H et al (2019) Novel and recurrent mutations in a cohort of chinese patients with young-onset amyotrophic lateral sclerosis. Front Neurosci 13:1289. https://doi.org/10.3389/fnins.2019.01289
Demontis F, Perrimon N (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143(5):813–825. https://doi.org/10.1016/j.cell.2010.10.007
McGuire SE, Mao Z, Davis RL (2004) Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE 2004(220):l6
Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA (1995) A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol 171(1):169–181
Deng J, Yang M, Chen Y, Chen X, Liu J, Sun S, Cheng H, Li Y et al (2015) FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet 11(9):e1005357. https://doi.org/10.1371/journal.pgen.1005357
Li Y, Ray P, Rao EJ, Shi C, Guo W, Chen X, Woodruff EA, Fushimi K et al (2010) A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci U S A 107(7):3169–3174. https://doi.org/10.1073/pnas.0913602107
Viscarra JA, Wang Y, Nguyen HP, Choi YG, Sul HS (2020) Histone demethylase JMJD1C is phosphorylated by mTOR to activate de novo lipogenesis. Nat Commun 11(1):796. https://doi.org/10.1038/s41467-020-14617-1
Zhang D, Zhai X, He X-j (2015) Application of oil red O staining in spinal cord injury of rats. Zhongguo Gu Shang 28(8):738–742
Deng J, Yu J, Li P, Luan X, Cao L, Zhao J, Yu M, Zhang W et al (2020) Expansion of GGC repeat in GIPC1 is associated with oculopharyngodistal myopathy. Am J Hum Genet 106(6):793–804. https://doi.org/10.1016/j.ajhg.2020.04.011
Cykowski MD, Powell SZ, Appel JW, Arumanayagam AS, Rivera AL, Appel SH (2018) Phosphorylated TDP-43 (pTDP-43) aggregates in the axial skeletal muscle of patients with sporadic and familial amyotrophic lateral sclerosis. Acta Neuropathol Commun 6(1):28. https://doi.org/10.1186/s40478-018-0528-y
Dupuis L, Gonzalez de Aguilar J-L, Echaniz-Laguna A, Eschbach J, Rene F, Oudart H, Halter B, Huze C et al (2009) Muscle mitochondrial uncoupling dismantles neuromuscular junction and triggers distal degeneration of motor neurons. PLoS One 4(4):e5390. https://doi.org/10.1371/journal.pone.0005390
Wong M, Martin LJ (2010) Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet 19(11):2284–2302. https://doi.org/10.1093/hmg/ddq106
Staunton L, Jockusch H, Ohlendieck K (2011) Proteomic analysis of muscle affected by motor neuron degeneration: the wobbler mouse model of amyotrophic lateral sclerosis. Biochem Biophys Res Commun 406(4):595–600. https://doi.org/10.1016/j.bbrc.2011.02.099
Chiang P-M, Ling J, Jeong YH, Price DL, Aja SM, Wong PC (2010) Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A 107(37):16320–16324. https://doi.org/10.1073/pnas.1002176107
Klivenyi P, Ferrante RJ, Matthews RT, Bogdanov MB, Klein AM, Andreassen OA, Mueller G, Wermer M et al (1999) Neuroprotective effects of creatine in a transgenic animal model of amyotrophic lateral sclerosis. Nat Med 5(3):347–350. https://doi.org/10.1038/6568
Kira Y, Nishikawa M, Ochi A, Sato E, Inoue M (2006) L-carnitine suppresses the onset of neuromuscular degeneration and increases the life span of mice with familial amyotrophic lateral sclerosis. Brain Res 1070(1):206–214. https://doi.org/10.1016/j.brainres.2005.11.052
Ivanova MI, Sievers SA, Guenther EL, Johnson LM, Winkler DD, Galaleldeen A, Sawaya MR, Hart PJ et al (2014) Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc Natl Acad Sci U S A 111 (1):197-201. doi:https://doi.org/10.1073/pnas.1320786110
Verdin E, Hirschey MD, Finley LWS, Haigis MC (2010) Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem Sci 35(12):669–675. https://doi.org/10.1016/j.tibs.2010.07.003
Bhat AH, Dar KB, Anees S, Zargar MA, Masood A, Sofi MA, Ganie SA (2015) Oxidative stress, mitochondrial dysfunction and neurodegenerative diseases; a mechanistic insight. Biomed Pharmacother 74:101–110. https://doi.org/10.1016/j.biopha.2015.07.025
Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J et al (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323(5918):1205–1208. https://doi.org/10.1126/science.1166066
Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J et al (2002) Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS). Proc Natl Acad Sci U S A 99(3):1604–1609. https://doi.org/10.1073/pnas.032539299
Ruffoli R, Bartalucci A, Frati A, Fornai F (2015) Ultrastructural studies of ALS mitochondria connect altered function and permeability with defects of mitophagy and mitochondriogenesis. Front Cell Neurosci 9:341. https://doi.org/10.3389/fncel.2015.00341
Golpich M, Amini E, Mohamed Z, Azman Ali R, Mohamed Ibrahim N, Ahmadiani A (2017) Mitochondrial dysfunction and biogenesis in neurodegenerative diseases: pathogenesis and treatment. CNS Neurosci Ther 23(1). https://doi.org/10.1111/cns.12655
Kodavati M, Wang H, Hegde ML (2020) Altered mitochondrial dynamics in motor neuron disease: an emerging perspective. Cells 9(4). https://doi.org/10.3390/cells9041065
Zhou J, Li A, Li X, Yi J (2019) Dysregulated mitochondrial Ca and ROS signaling in skeletal muscle of ALS mouse model. Arch Biochem Biophys 663:249–258. https://doi.org/10.1016/j.abb.2019.01.024
Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, DiPietro L, Cline GW et al (2003) Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science 300(5622):1140–1142. https://doi.org/10.1126/science.1082889
Carrière A, Carmona M-C, Fernandez Y, Rigoulet M, Wenger RH, Pénicaud L, Casteilla L (2004) Mitochondrial reactive oxygen species control the transcription factor CHOP-10/GADD153 and adipocyte differentiation: a mechanism for hypoxia-dependent effect. J Biol Chem 279(39):40462–40469. https://doi.org/10.1074/jbc.M407258200
Pu J, Ha CW, Zhang S, Jung JP, Huh W-K, Liu P (2011) Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell 2(6):487–496. https://doi.org/10.1007/s13238-011-1061-y
Acknowledgements
We thank the patients and their families for cooperation. We thank Mr. Lijun Chai and Mr. ** Xu (Peking University First Hospital) for their work for electron microscopy pictures, and Ms. Yuehuan Zuo and Ms. Qiurong Zhang (Peking University First Hospital) for preparations of pathological sections.
Funding
This study was supported by funding from the National Natural Science Foundation of China (Grant Nos. 81371394, 81460199, 81540101, 31701004, and 82160252), Natural Science Foundation of Jiangxi province (20202BAB206029), Double thousand talents program of Jiangxi province (jxsq2019101021).
Author information
Authors and Affiliations
Contributions
ZB, DJ, LX, ZY, DH, ZY, ZM, and YY contributed to the acquisition and analysis of data. FX, ZM, and WZ performed the genetic analysis. YJ, ZW, and YY performed the pathological study. YY and WZ contributed to critical revision of the manuscript. ZB, DJ, and HD contributed to the study design and drafted the manuscript.
Corresponding authors
Ethics declarations
Ethics Approval
The protocol was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University and the First Hospital of Peking University. All patients obtained informed consent to participate in the study.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Supplemental Fig. S1.
Cytotoxicity of Ctr, Wt, K510Q assessed by CTG assay. Cells were treated with Ctr, Wt or K510Q adenovirus for 48h and cell viability determined using the CellTiter-Glo assay. Values shown are mean ± SD of a representative experiment performed in sextuplicate (****: p<0.0001). (PNG 29 kb)

Supplemental Fig. S2.
Expression of FUS-K510Q causes lipid droplets accumulation. A Oil Red O staining of C2C12 cells without OA treatment. B The lipid droplet ratio was calculated using Image-Pro Plus software (****: p<0.0001). (PNG 1527 kb)

Supplemental Fig. S3.
Expression of Ctr, Wt, K510Q causes disordered lipid metabolism. A Oil Red O staining of C2C12 cells under 0.25 mM OA treatment. B The lipid droplet ratio was measured and quantified using Image-Pro Plus software (**: p<0.01). (PNG 1096 kb)
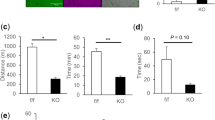
Expression of FUS-K510Q causes locomotor defects in flies at 35 days old. Fly genotypes: Ctr: Mef2-Gal4/Tub-Gal80ts/W1118; FUS-K510Q: Mef2-Gal4/Tub-Gal80ts/UAS-K510Q-FUS (MP4 1638 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhou, B., Zheng, Y., Li, X. et al. FUS Mutation Causes Disordered Lipid Metabolism in Skeletal Muscle Associated with ALS. Mol Neurobiol 59, 7265–7277 (2022). https://doi.org/10.1007/s12035-022-03048-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-022-03048-2