Abstract
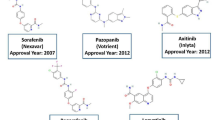
Renal cell carcinoma is a highly vascular tumor associated with vascular endothelial growth factor (VEGF) expression. The Vascular Endothelial Growth Factor -2 (VEGF-2) and its receptor was identified as a potential anti-cancer target, and it plays a crucial role in physiology as well as pathology. Inhibition of angiogenesis via blocking the signaling pathway is considered an attractive target. In the present study, 150 FDA-approved drugs have been screened using the concept of drug repurposing against VEGFR-2 by employing the molecular docking, molecular dynamics, grou** data with Machine Learning algorithms, and density functional theory (DFT) approaches. The identified compounds such as Pazopanib, Atogepant, Drosperinone, Revefenacin and Zanubrutinib shown the binding energy − 7.0 to − 9.5 kcal/mol against VEGF receptor in the molecular docking studies and have been observed as stable in the molecular dynamic simulations performed for the period of 500 ns. The MM/GBSA analysis shows that the value ranging from − 44.816 to − 82.582 kcal/mol. Harnessing the machine learning approaches revealed that clustering with K = 10 exhibits the relevance through high binding energy and satisfactory logP values, setting them apart from compounds in distinct clusters. Therefore, the identified compounds are found to be potential to inhibit the VEGFR-2 and the present study will be a benchmark to validate the compounds experimentally.


















Similar content being viewed by others
References
Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. The Lancet. 2009;373(9669):1119–32. https://doi.org/10.1016/s0140-6736(09)60229-4.
Motzer RJ, Rini BI, McDermott DF, Redman BG, Kuzel TM, Harrison MR, Vaishampayan UN, Drabkin HA, George S, Logan TF, Margolin KA, Plimack ER, Lambert AM, Waxman IM, Hammers HJ. Nivolumab for metastatic renal cell carcinoma: results of a randomized phase II trial. J Clin Oncol. 2015;33(13):1430–7. https://doi.org/10.1200/jco.2014.59.0703.
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Prime. 2017. https://doi.org/10.1038/nrdp.2017.9.
Broekman F. Tyrosine kinase inhibitors: multi-targeted or single-targeted? World J Clin Oncol. 2011;2(2):80. https://doi.org/10.5306/wjco.v2.i2.80.
Heng DY, **e W, Regan MM, Warren MA, Golshayan AR, Sahi C, Eigl BJ, Ruether JD, et al. Prognostic factors for overall survival in patients With metastatic renal cell carcinoma treated with vascular endothelial growth factor–targeted agents: results from a large, multicenter study. J Clin Oncol. 2009;27(34):5794–9. https://doi.org/10.1200/jco.2008.21.4809.
Hutson TE, Davis ID, Machiels JH, de Souza PL, Baker K, Bordogna W, Westlund R, Crofts T, Pandite L, Figlin RA. Biomarker analysis and final efficacy and safety results of a phase II renal cell carcinoma trial with pazopanib (GW786034), a multi-kinase angiogenesis inhibitor. J Clin Oncol. 2008;26:5046–504. https://doi.org/10.1200/jco.2008.26.15_suppl.5046.
Hasinoff BB, Patel D. The lack of target specificity of small molecule anticancer kinase inhibitors is correlated with their ability to damage myocytes in vitro. Toxicol Appl Pharmacol. 2010;249(2):132–9. https://doi.org/10.1016/j.taap.2010.08.026.
Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2018;18(1):41–58. https://doi.org/10.1038/nrd.2018.168.
Sliwoski G, Kothiwale S, Meiler J, Lowe EW. Computational methods in drug discovery. Pharmacol Rev. 2023;66(1):334–95. https://doi.org/10.1124/pr.112.007336.
Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26(1):80–93. https://doi.org/10.1016/j.drudis.2020.10.010.
Johnston RC, Yao K, Kaplan Z, Chelliah M, Leswing K, Seekins S, Watts S, Calkins D, et al. Epik: pKa and protonation state prediction through machine learning. J Chem Theory Comput. 2023;19:2380–8.
Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31(2):455–61. https://doi.org/10.1002/jcc.21334.
Berahmand K, Mohammadi M, Faroughi A, Mohammadiani RP. A novel method of spectral clustering in attributed networks by constructing a parameter-free affinity matrix. Cluster Comput. 2021;25(2):869–88. https://doi.org/10.1007/s10586-021-03430-0.
**ong G, Wu Z, Yi J, Fu L, Yang Z, Hsieh C, Yin M, Zeng X, Wu C, Lu A, Chen X, Hou T, Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49(W1):W5–14. https://doi.org/10.1093/nar/gkab255.
Borlea ID, Precup RE, Borlea AB. Improvement of K-means cluster quality by post processing resulted clusters. Procedia Comput Sci. 2022;199:63–70. https://doi.org/10.1016/j.procs.2022.01.009.
Miyamoto S, Kollman PA. Settle an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13(8):952–62. https://doi.org/10.1002/jcc.540130805.
Sathyamoorthy M, Kuppusamy S, Dhanaraj RK, Ravi V. Improved k-means based q learning- ing algorithm for optimal clustering and node balancing in wsn. Wireless Pers Commun. 2022;122(3):2745–66.
Wen J, Zhang Z, Fei L, Zhang B, Xu Y, Zhang Z, Li J. A survey on incomplete multiview clustering. IEEE Trans Syst Man Cybern. 2023;53(2):1136–49. https://doi.org/10.1109/tsmc.2022.3192635.
Abdullah D, Susilo S, Ahmar AS, Rusli R, Hidayat R. The application of K-means clustering for province clustering in Indonesia of the risk of the COVID-19 pandemic based on COVID-19 data. Quality Quantity. 2021;56(3):1283–91. https://doi.org/10.1007/s11135-021-01176-w.
Li T, Rezaeipanah A, Tag El Din EM. An ensemble agglomerative hierarchical clustering algorithm based on clusters clustering technique and the novel similarity measurement. J King Saud University-Comput Inform Sci. 2022;34(6):3828–42. https://doi.org/10.1016/j.jksuci.2022.04.010.
Eminagaoglu M, Oskay RG, Karayigit AI. Evaluation of elemental affinities in coal using agglomerative hierarchical clustering algorithm: a case study in a thick and mineable coal seam (kM2) from Soma Basin (W Turkey). Int J Coal Geol. 2022;259:104045. https://doi.org/10.1016/j.coal.2022.104045.
Wang Y, Ding S, Wang L, Du S. A manifold p-spectral clustering with sparrow search algorithm. Soft Comput. 2022;26(4):1765–77. https://doi.org/10.1007/s00500-022-06741-5
Srivastava PR, Sarkar P, Hanasusanto GA. A robust spectral clustering algorithm for Sub-Gaussian mixture models with outliers. Operations Res. 2023;71(1):224–44. https://doi.org/10.1287/opre.2022.2317.
Danielsson PE. Euclidean distance map**. Comput Gr Image Process. 1980;14(3):227–48. https://doi.org/10.1016/0146-664x(80)90054-4.
Wolber G, Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J Chem Inf Model. 2005;45(1):160–9. https://doi.org/10.1021/ci049885e.
Al-Asri J, Fazekas E, Lehoczki G, Perdih A, Görick C, Melzig MF, et al. From carbohydrates to drug-like fragments: Rational development of novel α-amylase inhibitors. Bioorg Med Chem. 2015;23(20):6725–32. https://doi.org/10.1016/j.bmc.2015.09.007.
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18(12):1463–72.
Soares TA, Daura X, Oostenbrink C, Smith LJ, van Gunsteren WF. Validation of the GROMOS force-field parameter set 45A3 against nuclear magnetic resonance data of hen egg lysozyme. J Biomol NMR. 2004;30(4):407–22. https://doi.org/10.1007/s10858-004-5430-1.
Gaussian 16, Revision C.01, Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Petersson GA, Nakatsuji H, Li X, Caricato M, Marenich AV, Bloino J, Janesko BG, Gomperts R, Mennucci B, Hratchian HP, Ortiz JV, Izmaylov JF, Sonnenberg JL, Williams-Young D, Ding F, Lipparini F, Egidi F, Goings J, Peng B, Petrone A, Henderson T, Ranasinghe D, Zakrzewski VG, Gao J, Rega N, Zheng G, Liang W, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Throssell K, Montgomery JA, Peralta JE, Ogliaro F, Bearpark MJ, Heyd JJ, Brothers EN, Kudin KN, Staroverov VN, Keith TA, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Millam JM, Klene M, Adamo C, Cammi R, Ochterski JW, Martin RL, Morokuma K, Farkas O, Foresman JB, Fox DJ. Gaussian, Inc., Wallingford CT, 2016.
Balajee R, Srinivasadesikan V, Sakthivadivel M, Gunasekaran P. In SilicoScreening, alanine mutation, and DFT approaches for identification of NS2B/NS3 protease inhibitors. Biochem Res Int. 2016;2016:1–13. https://doi.org/10.1155/2016/7264080.
Acknowledgements
The authors VSD, VSJ and SS acknowledge VFSTR for providing Computational support to carry out this work.
Funding
Nil.
Author information
Authors and Affiliations
Contributions
VSJ, VSD: Conceptualization, design of work. VSJ, BR, VSD: Data curation, visualization, validation, writing—original draft. VSJ, BR, SS, VSD: Molecular Docking. BR, RM: Molecular Dynamics Simulations. ACB, BR: Machine Learning concepts. Supervision and Investigation: VSD, BR. The final version of the manuscript submitted was approved by all the authors.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the author(s).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jerra, V.S., Ramachandran, B., Shareef, S. et al. Molecular docking aided machine learning for the identification of potential VEGFR inhibitors against renal cell carcinoma. Med Oncol 41, 198 (2024). https://doi.org/10.1007/s12032-024-02419-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-024-02419-0