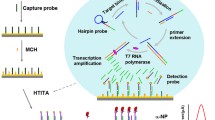
Abstract
Early determination of infectious pathogens is vitally important to select appropriate antibiotics, and to manage nosocomial infection. Herein, we propose a target recognition triggered triple signal amplification–based approach for sensitive pathogenic bacteria detection. In the proposed approach, a double-strand DNA probe (capture probe) that is composed of an aptamer sequence and a primer sequence is designed for specific identification of target bacteria and initiation of following triple signal amplification. After recognition of target bacteria, primer sequence is released from capture probe to bind with the designed H1 probe, forming a blunt terminal in the H1 probe. Exonuclease-III (Exo-III enzyme) specifically recognizes the blunt terminal in H1 probe and degrades the sequence from 3′ terminal, resulting a single-strand DNA to induce the following signal amplification. Eventually, the approach exhibits a low detection limit of 36 cfu/mL with a broad dynamic range. The high selectivity endows the method a promising prospective for clinical sample analysis.
Graphical abstract





Similar content being viewed by others
Data Availability
All authors have confirmed that the data used in this manuscript have been presented clearly, honestly, and without falsification or inappropriate data manipulation (including image-based manipulation).
Code Availability
Not applicable.
References
Vasquez, V., Ampuero, D., & Padilla, B. (2017). Urinary tract infections in inpatients: That challenge. Revista Española de Quimioterapia, 30(Suppl 1), 39–41.
Foxman, B. (2010). The epidemiology of urinary tract infection. Nature Reviews Urology, 7, 653–660.
Kalinderi, K., Delkos, D., Kalinderis, M., Athanasiadis, A., & Kalogiannidis, I. (2018). Urinary tract infection during pregnancy: Current concepts on a common multifaceted problem. Journal of Obstetrics and Gynaecology, 38, 448–453.
Salvatore, S., Salvatore, S., Cattoni, E., Siesto, G., Serati, M., Sorice, P., & Torella, M. (2011). Urinary tract infections in women. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 156, 131–136.
Gokce, S. and Kurugol, Z (2020) Diagnostic accuracy of the mean platelet volume in the prediction of upper urinary tract infections. Clin Lab, 66, 190647.
Beveridge, L. A., Davey, P. G., Phillips, G., & McMurdo, M. E. (2011). Optimal management of urinary tract infections in older people. Clinical Interventions in Aging, 6, 173–180.
Leitner, L., Ujmajuridze, A., Chanishvili, N., Goderdzishvili, M., Chkonia, I., Rigvava, S., Chkhotua, A., Changashvili, G., McCallin, S., Schneider, M. P., Liechti, M. D., Mehnert, U., Bachmann, L. M., Sybesma, W., & Kessler, T. M. (2021). Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomised, placebo-controlled, double-blind clinical trial. The Lancet Infectious Diseases, 21, 427–436.
Pala L, Sirec T and Spitz U (2020) Modified enzyme substrates for the detection of bacteria: A review. Molecules, 25, 3690.
Jones, L. M., Dunham, D., Rennie, M. Y., Kirman, J., Lopez, A. J., Keim, K. C., Little, W., Gomez, A., Bourke, J., Ng, H., DaCosta, R. S., & Smith, A. C. (2020). In vitro detection of porphyrin-producing wound bacteria with real-time fluorescence imaging. Future Microbiology, 15, 319–332.
Li, D., Kumari, B., Makabenta, J. M., Gupta, A., & Rotello, V. (2019). Effective detection of bacteria using metal nanoclusters. Nanoscale, 11, 22172–22181.
Lu, L., Liu, M., Sun, R., Zheng, Y., & Zhang, P. (2015). Myocardial infarction: Symptoms and treatments. Cell Biochemistry and Biophysics, 72, 865–867.
Lagier, J. C., Edouard, S., Pagnier, I., Mediannikov, O., Drancourt, M., & Raoult, D. (2015). Current and past strategies for bacterial culture in clinical microbiology. Clinical Microbiology Reviews, 28, 208–236.
Rajapaksha, P., Elbourne, A., Gangadoo, S., Brown, R., Cozzolino, D., & Chapman, J. (2019). A review of methods for the detection of pathogenic microorganisms. The Analyst, 144, 396–411.
Liu, Y., Zhou, H., Hu, Z., Yu, G., Yang, D., & Zhao, J. (2017). Label and label-free based surface-enhanced Raman scattering for pathogen bacteria detection: A review. Biosensors & Bioelectronics, 94, 131–140.
Li, Y., Fan, P., Zhou, S., & Zhang, L. (2017). Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microbial Pathogenesis, 107, 54–61.
Li, D., Liu, L., Huang, Q., Tong, T., Zhou, Y., Li, Z., Bai, Q., Liang, H., & Chen, L. (2021). Recent advances on aptamer-based biosensors for detection of pathogenic bacteria. World Journal of Microbiology & Biotechnology, 37, 45.
Zhao, W., Wang, L., & Tan, W. (2007). Fluorescent nanoparticle for bacteria and DNA detection. Advances in Experimental Medicine and Biology, 620, 129–135.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Jie Guo and Qun Liang coordinated all the experiments and wrote the manuscript draft. Huajun Zhang, Guo Wei, and Wantao Zhang designed the approach and revised the manuscript. Huifang Zhang and Miao Tian analyzed the research data and reviewed the manuscript. All authors have read and approved the final manuscript. Jie Guo, Qun Liang, and Huifang Zhang contributed equally to this work
Corresponding authors
Ethics declarations
Ethics Approval
This article does not contain any studies with human participants or animals.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guo, J., Liang, Q., Zhang, H. et al. Exo-III Enzyme–Assisted Triple Cycle Signal Amplifications for Sensitive and Accurate Identification of Pathogenic Bacteria. Appl Biochem Biotechnol 195, 6203–6211 (2023). https://doi.org/10.1007/s12010-023-04391-3
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04391-3