Abstract
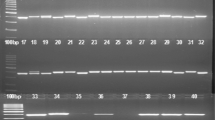
Throughout the globe morphological, biochemical and genetic variability exists in chilli and is harnessed to achieve specific breeding objectives. In this study, chilli germplasm was characterized based on horticultural traits, biochemical quantification and simple sequence repeat (SSR) polymorphism for diversity estimation. A total of 36 SSR primers were utilised to study the genetic divergence among 48 genotypes of chilli collected from nine states of India. Among the 36 primers, sixteen amplified null alleles. A total of 41 alleles were detected with average 2.05 alleles per locus. The largest number of alleles (5) were obtained with marker CAMS-234. The polymorphic information content ranged from 0.06 to 0.72 with an average of 0.50. On the basis of SSR analysis, the UPGMA cluster classified 48 genotypes into three groups. There was significant variability in germplasm for all morpho-biochemical traits. Kashi Anmol (100.50 q/ha) expressed the highest yield. Highest vitamin C content at green stage was recorded in IC-561635 (187 mg/100 g) and the greatest capsaicin content (9547.90 µg/g) equivalent to pungency of 171,862.2 Scoville heat units (SHU) was recorded in Bhut Jolokia. Principal component analysis indicates that the first five principal components explain 74.63% per cent of the total variation. Additionally, analysis of molecular variance (AMOVA) showed that 1% of the total genetic variation occurred among the population and 99% genetic variation within the populations, whereas the pairwise Fst specified the moderate genetic variation ranging from 0.002 to 0.020. The present investigation has strengthened the knowledge of genetic worth of this germplasm for application in various genetic improvement programmes.







Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Ahmed N, Hou X (2022) Genetic variation and population structure analysis of Leymus chinensis (Trin.) Tzvelev from Eurasian steppes using SSR makers. Genet Res Crop Evol 69:2425–2436
AOAC (1984) Official Methods of Analysis. Association of official analytical chemists 13th edition, Washington D.C. pp 497
Aremu CO (2011) Genetic diversity: a review for need and measurements for intraspecies crop improvement. J Microbiol Biotechol Res 1(2):80–85
ASTA (1986) Official analytical methods of the American Spice Trade Association, 2nd edn. ASTA, Englewood Chiffs, N. I.
Bhattacharya A, Chattopadhyay A, Mazumdar D, Chakravarty A, Pal S (2010) Antioxidant constituents and enzymes activities in chilli peppers. Int J Veg Sci 16(3):201–211
Bosland PW, Baral JB (2007) Bhut Jolokia- the World’s hottest known chile pepper is a putative naturally occurring interspecific hybrid. Hort Sci 30:137–139
Bosland PW, Votava EJ (2003) Peppers: vegetable and spice Capsicum. CAB International, England, p 333
Costa FRD, Pereira TNS, Vitoria AP, De Campos KP, Rodrigues R, Da Silva DH, Pereira MG (2006) Genetic diversity among Capsicum accessions using RAPD markers. Crop Breed Appl Biotechnol 6:18–23
Dan M (2012) The red-hot chilli cook book: fabulously fiery recipes for chilli fans, published by Ryland Peters and small 20–21 Jockey’s fields London WCIR 4 BW and 519 Broadway, New York
Dewitt D (1999) The Chile Pepper Encyclopedia. New Work, William Marrow
Dhaliwal MS, Yadav A, **dal SK (2014) Molecular characterization and diversity analysis in chilli pepper using simple sequence repeats (SSR) markers. African J Biotechnol 13(31):3137–3143
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12(1):13–15
Farhad M, Hasanuzzaman M, Biswas BK, Arifuzzaman M, Islam MM (2010) Genetic divergence in chilli (Capsicum annuum L.). Bangladesh Res Pub J 3(3):1045–1051
Fonseca RM, Lopes R, Barros SW, Lopes MTG, Ferreira FM (2008) Morphological characterization and genetic diversity of Capsicum chinensis Jacq accessions along the upper Rio-Negro-Amazonas. Crop Breed Appl Biotechnol 8:187–194
Geleta LF, Labuschagne MT, Viljoen CD (2005) Genetic variability in pepper (Capsicum annuum L.) estimated by morphological data and amplified fragment length polymorphism markers. Biodivers Conserv 14:2361–2375
Ghosh A, Pugalendhi L (2012) Studies on the improvement of chilli (Capsicum annuum L.) cv. Co 4 for quality characters through hybridization. Elec J Plant Breed 3(2):811–817
Gilbert JE, Lewis RV, Wilkinson MJ, Caligari PDF (1999) Develo** an appropriate strategy to assess genetic variability in plant germplasm collection. Theor Appl Genet 98:1125–1131
Harvell KP, Bosland PW (1997) The environment produces a significant effect on pungency of chiles (Capsicum annuum L.). Hort Sci 32:1292
Hasanuzzaman M, Golam F (2011) Selection of parents in chilli (Capsicum annuum L.). Int J Sus Crop Prod 6(1):63–75
IPGRI (1999) Isolated from a size-fractionated genomic library of Brassica napus L. (Rapeseed). Theor Appl Genet 91:206–211
Janaki M, Naidu LN, Ramana CV, Rao MP (2016) Genetic divergence among chilli (Capsicum annuum L.) genotypes based on quantitative and qualitative traits. Int J Sci Nat 7(1):181–189
Karad SR, Raikar GR, Navale PA (2002) Genetic divergence in chilli. J Maharashtra Agric Univ 27(2):143–145
Khodadabi M, Fotokian MH, Miransari M (2011) Genetic diversity of wheat genotypes based on cluster and principal component analysis for breeding strategies. Aust J Crop Sci 5(1):17–24
Kumar B, Lal G, Ruchi M, Upadhyay A (2009) Genetic variability, diversity and association of quantitative traits with grain yield in bread wheat (Triticum aestivum L.). Asian J Agri Sci 1(1):4–6
Kumar BMD, Kantti A, Mallikarjunaiah H (2010) Genetic divergence in chilli accessions. Electronic J Plant Breeding 1(5):1363–1366
Kwon YS, Lee JM, Yi GB, Yi SI, Kim KM, Soh EH (2005) Use of SSR markers to compliment tests of distinctiveness, uniformity and stability (DUS) of pepper (Capsicum annuum L. var. annuum) varieties. Mol Cells 19:428–435
Lannes SD, Finger FL, Schuelter AR, Casali VWD (2007) Growth and quality of Brazilian accessions of Capsicum chinense fruits. Sci Hortic 112:266–270
Laurentin H (2009) Data analysis for molecular characterization of plant genetic resources. Genet Res Crop Evol 56:277–292
Mahalanobis PC (1936) On the generalized distance in statistics. Proc Nat Inst Sci Indi 2:49–55
Minamiyama Y, Tsuro M, Hirai M (2006) An SSR based linkage map of Capsicum annuum. Mol Breed 18:157–169
Nahak SC, Nandi A, Sahu GS, Tripathy P, Das S, Mohanty A, Pradhan SR (2018) Assessment of genetic diversity in different chilli (Capsicum annuum L.) genotypes. Int J Curr Microbiol Appl Sci 7(9):634–639
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of Restriction endonucleases. Proc Natl Acad Sci USA 79:5269–5273
Paran I, Knaap EV (2007) Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot 58(14):3841–3852
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. population genetic software for teaching and research- an update. Bioinformatics 28:2537–2539
Perry L, Dickau R, Zarrillo S, Holst I, Pearsall DM, Piperno DR, Berman MJ, Cooke RG, Rademaker K, Ranere AJ, Rademaker K, Ranere AJ, Raymond JS, Sandweiss DH, Scaramelli F, Tarble K, Zeidler JA (2007) Starch fossils and the domestication and dispersal of chilli pepper (Capsicum spp. L.). Americas Science 315:986–998
Pradhan K, Nandi A, Das A, Sahu, Senapati N (2018) Quantification of capsaicin and ascorbic acid content in twenty-four Indian genotypes of chilli (Capsicum annuum L.) by HPTLC and Volumetric method. Int J Pure Appl Bioscience 6(1):1322–1327
Pritchard JK, Stephens M (2000) Inference of population structure using multilocus genotype data. Genetics 155(945–59):56
Rao CR (1952) Advanced statistical methods in biometrical research. Wiley, New York
Rohlf FJ (1997) NTSYS-PC 2.1: numerical taxonomy and multivariate analysis system. Setauket Newyork Exeter software
Rymbai H, Sharma RR, Srivastav M (2011) Bio-colorants and its implications in health and food industry—a review. Int J Pharm Tech Res 3:2228–2244
Salvador MH (2002) Genetic resources of chilli (Capsicum annuum L.) in Mexico. In: Proceedings of the 16th International Pepper Conference, Tampico, Tamaulipas, Mexico. pp 10–12
Sharmin A, Hoque ME, Haque MdM, Khatun F (2018) Molecular diversity analysis of some chilli (Capsicum spp.) genotypes using SSR markers. Amer J Plant Sci 9:368–379
Singh RK, Sharma RK, Singh AK, Singh VP, Singh NK, Tiwari SP, Mohapatra T (2004) Suitability of mapped sequence tagged microsatellite site markers for establishing distinctness, uniformity and stability in aromatic rice. Euphytica 135:135–143
Srinivas B, Thomas B, Gogineni S (2013) Genetic divergence for yield and its component traits in chilli (Capsicum frutescens L.) accessions of Kerala. Int J Sci Res 4:2319–7064
Tanksley SD (1984) High rates of cross pollination in chilli pepper. Hort Sci 4:580–582
Wang J, Yao J, Li W (2008) Construction of a molecular map for melon (Cucumis melo L.) based on SRAP. Front Agric China 2(4):451–455
Ye-yun X, Zhan Z, Yi-** X, Long- ** Y (2005) Identification and purity test of super hybrid rice with SSR molecular markers. Rice Sci 12(1):7–12
Yuzbapyoglu E, Ozcan S, Acik L (2006) Analysis of genetic relationships among Turkish cultivars and breeding lines of lens culinatis mestile using RAPD markers. Genet Res Crop Evol 53:507–514
Zehra SB (2014) Msc. thesis submitted to Division of Vegetable Science, SKUAST-Kashmir, Shalimar Srinagar
Zitter TA, Hopkins DL, Thomas CE (1996) Compendium of Cucurbit Diseases. APS Press, St. Paul, p 87
Acknowledgements
This work was supported by grant provided by all India coordinated research project on vegetable crops (AICRP-VC), research grant (Division of vegetable science SKUAST-K) and this research was funded by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (2019R1A6A1A11052070).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Indrabi, S.A., Malik, A., Malik, G. et al. Phenotypic, biochemical and genetic diversity of pepper (Capsicum spp.) germplasm reflects selection for cultivar types and spatial distribution. Plant Biotechnol Rep 18, 341–360 (2024). https://doi.org/10.1007/s11816-024-00904-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-024-00904-6