Abstract
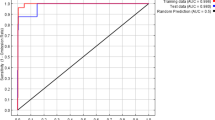
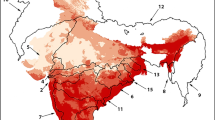
China has become one of the most serious countries suffering from biological invasions in the world. In the context of global climate change, invasive alien species (IAS) are likely to invade a wider area, posing greater ecological and economic threats in China. Western mosquitofish (Gambusia affinis), which is known as one of the 100 most invasive alien species, has distributed widely in southern China and is gradually spreading to the north, causing serious ecological damage and economic losses. However, its distribution in China is still unclear. Hence, there is an urgent need for a more convenient way to detect and monitor the distribution of G. affinis to put forward specific management. Therefore, we detected the distribution of G. affinis in China under current and future climate change by combing Maxent modeling prediction and eDNA verification, which is a more time-saving and reliable method to estimate the distribution of species. The Maxent modeling showed that G. affinis has a broad habitat suitability in China (especially in southern China) and would continue to spread in the future with ongoing climate change. However, eDNA monitoring showed that occurrences can already be detected in regions that Maxent still categorized as unsuitable. Besides temperature, precipitation and human influence were the most important environmental factors affecting the distribution of G. affinis in China. In addition, by environmental DNA analysis, we verified the presence of G. affinis predicted by Maxent in the Qinling Mountains where the presence of G. affinis had not been previously recorded.






Similar content being viewed by others
Data availability
All data is available in this article. The datasets generated and/or analyzed during the current study are available in the GenBank repository (LC193305.1) and from the corresponding author on reasonable request.
References
Bohan DA, Vacher C, Tamaddoni-Nezhad A, Raybould A, Dumbrell AJ, Woodward G (2017) Next-generation global biomonitoring: large-scale, automated reconstruction of ecological networks. Trends Ecol Evol 32:477–487. https://doi.org/10.1016/j.tree.2017.03.001
Brown JL, Bennett JR, French CM (2017) SDMtoolbox 2.0: the next generation Python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. PeerJ 5:e4095. https://doi.org/10.7717/peerj.4095
Chen D, Li D (1996) Development prospect of Channo argus pond culture. Jiangxi Aquacult Sci Technol 23:46–48
Cobos ME, Peterson AT, Barve N, Osorio-Olvera L (2019) kuenm: an R package for detailed development of ecological niche models using Maxent. PeerJ 7:e6281. https://doi.org/10.7717/peerj.6281
Courchamp F, Fournier A, Bellard C, Bonnaud E, Jeschke JM, Russell JC (2017) Invasion biology: specific problems and possible solutions. Trends Ecol Evol 32:13–22. https://doi.org/10.1016/j.tree.2016.11.001
Diagne C, Leroy B, Gozlan RE, Vaissière A-C, Assailly C, Nuninger L, Roiz D, Jourdain F, Jarić I, Courchamp F (2020) InvaCost, a public database of the economic costs of biological invasions worldwide. Sci Data 7:e277. https://doi.org/10.1038/s41597-020-00586-z
Dudgeon D, Corlett R (2011) The ecology and biodiversity of Hong Kong (revised edition). Agriculture, Fisheries and Conservation Department, Government of Hong Kong SAR and Joint Publishing Co, Hong Kong
Elith J, Graham CH, Anderson RP, Dudík M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton J, Peterson AT, Phillips SJ, Richardson KS, Scachetti-Pereira R, Schapire RE, Soberòn J, Williams S, Wisz MS, Zimmermann NE (2006) Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29:129–151. https://doi.org/10.1111/j.2006.0906-7590.04596.x
Ficetola GF, Poulenard J, Sabatier P, Messager E, Gielly L, Leloup A, Etienne D, Bakke J, Malet E, Fanget B, Storen E, Reyss JL, Taberlet P, Arnaud F (2018) DNA from lake sediments reveals long-term ecosystem changes after a biological invasion. Sci Adv 4:eaar4292. https://doi.org/10.1126/sciadv.aar4292
Fordham DA, Jackson ST, Brown SC, Huntley B, Brook BW, Dahl-Jensen D, Gilbert MTP, Otto-Bliesner BL, Svensson A, Theodoridis S, Wilmshurst JM, Buettel JC, Canteri E, McDowell M, Orlando L, Pilowsky J, Rahbek C, Nogues-Bravo D (2020) Using paleo-archives to safeguard biodiversity under climate change. Science 369:eabc5654. https://doi.org/10.1126/science.abc5654
Gao J, Ouyang X, Chen B, Jourdan J, Plath M (2017) Molecular and morphometric evidence for the widespread introduction of Western mosquitofish Gambusia affinis (Baird and Girard, 1853) into freshwaters of mainland China. BioInvasions Rec 6:281–289. https://doi.org/10.3391/bir.2017.6.3.14
García-Berthou E, Alcaraz C, Pou-Rovira Q, Zamora L, Coenders G, Feo C (2005) Introduction pathways and establishment rates of invasive aquatic species in Europe. Can J Fish Aquat Sci 62:453–463. https://doi.org/10.1139/f05-017
Guan X, Zhang J, Bao Z, Liu C, ** J, Wang G (2021) Past variations and future projection of runoff in typical basins in 10 water zones. China Sci Total Environ 798:149277. https://doi.org/10.1016/j.scitotenv.2021.149277
Henley SF, Schofield OM, Hendry KR, Schloss IR, Steinberg DK, Moffat C, Peck LS, Costa DP, Bakker DCE, Hughes C, Rozema PD, Ducklow HW, Abele D, Stefels J, Van Leeuwe MA, Brussaard CPD, Buma AGJ, Kohut J, Sahade R, Friedlaender AS, Stammerjohn SE, Venables HJ, Meredith MP (2019) Variability and change in the west Antarctic Peninsula marine system: research priorities and opportunities. Prog Oceanogr 173:208–237. https://doi.org/10.1016/j.pocean.2019.03.003
Huang G, Liu Y, Liang Y, Shi W, Yang Y, Liu S, Hu L, Chen H, **e L, Ying G (2019) Endocrine disrupting effects in western mosquitofish Gambusia affinis in two rivers impacted by untreated rural domestic wastewaters. Sci Total Environ 683:61–70. https://doi.org/10.1016/j.scitotenv.2019.05.231
Huang X, Liu H, Chen J, Gui L, Huang L (2022) Drivers of temporal variations in fish assemblages from mangrove creeks in Beihai, southern China. Environ Sci Pollut Res Int 29:47070–47081. https://doi.org/10.1007/s11356-022-19029-w
Hulme PE (2017) Climate change and biological invasions: evidence, expectations, and response options. Biol Rev Camb Philos Soc 92:1297–1313. https://doi.org/10.1111/brv.12282
Janosik A, Johnston C (2015) Environmental DNA as an effective tool for detection of imperiled fishes. Environ Biol Fish 98:1889–1893. https://doi.org/10.1007/s10641-015-0405-5
Jourdan J, Riesch R, Cunze S (2021) Off to new shores: climate niche expansion in invasive mosquitofish (Gambusia spp.). Ecol Evol 11:18369–18400. https://doi.org/10.1002/ece3.8427
Krumholz LA (1944) Northward acclimatization of the western mosquitofish, Gambusia affinis affinis. Copeia 1944:82–85
Krumholz LA (1948) Reproduction in the western mosquitofish, Gambusia affinis affinis (Baird & Girard), and its use in mosquito control. Ecol Monogr 18:1–43. https://doi.org/10.2307/1948627
Lan X, Li W, Tang J, Shakoor A, Zhao F, Fan J (2022) Spatiotemporal variation of climate of different flanks and elevations of the Qinling-Daba mountains in China during 1969–2018. Sci Rep 12:6952. https://doi.org/10.1038/s41598-022-10819-3
Li Z, **e Y (2002) Alien invasive species in China. China Forestry Publishing House, Bei**g, China
Lin Y, Gao Z, Zhan A (2015) Introduction and use of non-native species for aquaculture in China: status, risks and management solutions. Rev Aquac 7:28–58. https://doi.org/10.1111/raq.12052
Liu C, Diagne C, Angulo E, Banerjee A-K, Chen Y, Cuthbert RN, Haubrock PJ, Kirichenko N, Pattison Z, Watari Y, **ong W, Courchamp F (2021) Economic costs of biological invasions in Asia. NeoBiota 67:53–78. https://doi.org/10.3897/neobiota.67.58147
Lowe S, Browne M, Boudjelas S, De Poorter M (2000) 100 of the world’s worst invasive alien species. A selection from the global invasive species database. Invasive Species Specialist Group, Auckland, New Zealand
Lynggaard C, Bertelsen M, Jensen C, Johnson M, Frøslev T, Olsen M, Bohmann K (2022) Airborne environmental DNA for terrestrial vertebrate community monitoring. Curr Biol 32. https://doi.org/10.1016/j.cub.2021.12.014
Miklós B, Pfenninger M, Grossart HP, Taberlet P, Vellend M, Leibold MA, Englund G, Bowler D (2018) Environmental DNA time series in ecology. Trends Ecol Evol 33:945–957. https://doi.org/10.1016/j.tree.2018.09.003
Miya M, Sato Y, Fukunaga T, Sado T, Poulsen JY, Sato K, Minamoto T, Yamamoto S, Yamanaka H, Araki H, Kondoh M, Iwasaki W (2015) MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: detection of more than 230 subtropical marine species. R Soc Open Sci 2:150088. https://doi.org/10.1098/rsos.150088
Moorhouse T, Macdonald D (2015) Are invasives worse in freshwater than terrestrial ecosystems? Wires Water 2:1–8. https://doi.org/10.1002/wat2.1059
Murphy C, Gaël G, García-Berthou E (2015) Natural abiotic factors more than anthropogenic perturbation shape the invasion of Eastern Mosquitofish (Gambusia holbrooki). Freshw Sci 34:965–974. https://doi.org/10.1086/681948
Neto S, Sutton WB, Spear SF, Freake MJ, Kéry M, Schmidt BR (2020) Integrating species distribution and occupancy modeling to study hellbender (Cryptobranchus alleganiensis) occurrence based on eDNA surveys. Biol Conserv. https://doi.org/10.1016/j.biocon.2020.108787
Otto RG (1973) Temperature tolerance of the mosquitofish, Gambmia affinis (Baird and Girard). J Fish Biol 5:575–585. https://doi.org/10.1111/j.1095-8649.1973.tb04490.x
Ouyang X, Gao J, **e M, Liu B, Zhou L, Chen B, Jourdan J, Riesch R, Plath M (2018) Natural and sexual selection drive multivariate phenotypic divergence along climatic gradients in an invasive fish. Sci Rep 8:11164. https://doi.org/10.1038/s41598-018-29254-4
Pei H, Liu M, Shen Y, Xu K, Zhang H, Li Y, Luo J (2021) Quantifying impacts of climate dynamics and land-use changes on water yield service in the agro-pastoral ecotone of northern China. Sci Total Environ 809:151153. https://doi.org/10.1016/j.scitotenv.2021.151153
Phillips SJ, Dudík M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175. https://doi.org/10.1111/j.0906-7590.2008.5203.x
Purcell K, Hitch A, Klerks P, Leberg P (2008) Adaptation as a potential response to sea-level rise: a genetic basis for salinity tolerance in populations of a coastal marsh fish. Evol Appl 1:155–160. https://doi.org/10.1111/j.1752-4571.2007.00001.x
Pyke G (2005) A review of the biology of Gambusia affinis and G. holbrooki. Rev Fish Biol Fisher 15:339–365. https://doi.org/10.1007/s11160-006-6394-x
Qin Z, Zhang J, DiTommaso A, Wang R, Wu R (2015) Predicting invasions of Wedelia trilobata (L.) Hitchc. with Maxent and GARP models. J Plant Res 128. https://doi.org/10.1007/s10265-015-0738-3
Seebens H, Blackburn TM, Dyer EE, Genovesi P, Hulme PE, Jeschke JM, Pagad S, Pysek P, van Kleunen M, Winter M, Ansong M, Arianoutsou M, Bacher S, Blasius B, Brockerhoff EG, Brundu G, Capinha C, Causton CE, Celesti-Grapow L, Dawson W, Dullinger S, Economo EP, Fuentes N, Guenard B, Jager H, Kartesz J, Kenis M, Kuhn I, Lenzner B, Liebhold AM, Mosena A, Moser D, Nentwig W, Nishino M, Pearman D, Pergl J, Rabitsch W, Rojas-Sandoval J, Roques A, Rorke S, Rossinelli S, Roy HE, Scalera R, Schindler S, Stajerova K, Tokarska-Guzik B, Walker K, Ward DF, Yamanaka T, Essl F (2018) Global rise in emerging alien species results from increased accessibility of new source pools. Proc Natl Acad Sci USA 115:E2264–E2273. https://doi.org/10.1073/pnas.1719429115
Shi X, Chen X, Dai Y, Hu G (2020) Climate sensitivity and feedbacks of BCC-CSM to idealized CO2 forcing from CMIP5 to CMIP6. J Meteorol Res 34:865–878. https://doi.org/10.1007/s13351-020-9204-9
Tsang AHF, Dudgeon D (2021a) A manipulative field experiment reveals the ecological effects of invasive mosquitofish (Gambusia affinis) in a tropical wetland. Freshwater Biol 66:869–883. https://doi.org/10.1111/fwb.13683
Tsang AHF, Dudgeon D (2021b) Do exotic poeciliids affect the distribution or trophic niche of native fishes? Absence of evidence from Hong Kong streams. Freshwater Biol 66:1751–1764. https://doi.org/10.1111/fwb.13789
Wan F, Yang N (2016) Invasion and management of agricultural alien insects in China. Annu Rev Entomol 61:77–98. https://doi.org/10.1146/annurev-ento-010715-023916
Wang X, Gao T, Mo Z, Chen X (2017) Freshwater fish diversity in Weizhou Island. Guangxi Sciences 24:504–508. https://doi.org/10.13656/j.cnki.gxkx.20170921.001
Wang Y, Liu Y, Ma M, Ding Z, Wu S, Jia W, Chen Q, Yi X, Zhang J, Li X, Luo G, Huang J (2022) Dam-induced difference of invasive plant species distribution along the riparian habitats. Sci Total Environ 808:152103. https://doi.org/10.1016/j.scitotenv.2021.152103
Wildlife Conservation Society, Center for International Earth Science Information Network of Columbia University (2005) Last of the Wild Project, Version 2, 2005 (LWP-2): Global Human Footprint Dataset (Geographic). NASA Socioeconomic Data and Applications Center, Palisades, New York. https://doi.org/10.7927/H4M61H5F
Wu T, Lu Y, Fang Y, **n X, Li L, Li W, Jie W, Zhang J, Liu Y, Zhang L, Zhang F, Zhang Y, Wu F, Li J, Chu M, Wang Z, Shi X, Liu X, Wei M, Huang A, Zhang Y, Liu X (2019) The Bei**g Climate Center Climate System Model (BCC-CSM): the main progress from CMIP5 to CMIP6. Geosci Model Dev 12:1573–1600. https://doi.org/10.5194/gmd-12-1573-2019
**e Y, Li ZY, Gregg WP, Li DM (2000) Invasive species in China - an overview. Biodivers Conserv 10:1317–1341. https://doi.org/10.1023/A:1016695609745
Yang X, Zhou B, Xu Y, Han Z (2021) CMIP6 evaluation and projection of temperature and precipitation over China. Adv Atmos Sci 38:817–830. https://doi.org/10.1007/s00376-021-0351-4
Yin Y, He Q, Pan X, Liu Q, Wu Y, Li X (2022) Predicting current potential distribution and the range dynamics of Pomacea canaliculata in China under Global Climate Change. Biology 11:110. https://doi.org/10.3390/biology11010110
Zhang P, Dong X, Grenouillet G, Lek S, Zheng Y, Chang J (2020) Species range shifts in response to climate change and human pressure for the world’s largest amphibian. Sci Total Environ 735:139543. https://doi.org/10.1016/j.scitotenv.2020.139543
Zhao Z, **ao N, Shen M, Li J (2022) Comparison between optimized MaxEnt and random forest modeling in predicting potential distribution: a case study with Quasipaa boulengeri in China. Sci Total Environ 842:156867. https://doi.org/10.1016/j.scitotenv.2022.156867
Zhou L, Liu K, Zhao Y, Cui L, Dong C, Wang Z (2022a) Increasing salinization of freshwater limits invasiveness of a live-bearing fish: insights from behavioral and life-history traits. Environ Pollut 308:119658. https://doi.org/10.1016/j.envpol.2022.119658
Zhou L, Ouyang X, Zhao Y, Gomes-Silva G, Segura-Munoz SI, Jourdan J, Riesch R, Plath M (2022b) Invasive fish retain plasticity of naturally selected, but diverge in sexually selected traits. Sci Total Environ 811:152386. https://doi.org/10.1016/j.scitotenv.2021.152386
Zhuang H, Zhang C, ** X, Ge A, Chen M, Ye J, Qiao H, **ong P, Zhang X, Chen J, Luan X, Wang W (2022) A flagship species-based approach to efficient, cost-effective biodiversity conservation in the Qinling Mountains. China J Environ Manage 305:114388. https://doi.org/10.1016/j.jenvman.2021.114388
Acknowledgements
We are deeply thankful to the World Climate Research Program, Global Biodiversity Information Facility, WorldClim, NASA Socioeconomic Data and Applications Center, and Geospatial Data Cloud for the data we used. Special thanks to Fenzhi Lu for technical assistance, Yu Zhao, Kui Tang, Shuai Zhang, He Wang, and Xuening Li for their assistance in the field and in the lab. We also acknowledge the anonymous reviewers for comments of the manuscript.
Funding
This work was supported by grant from the National Natural Science Foundation of China for Youth (32001196).
Author information
Authors and Affiliations
Contributions
Conceptualization: Xu Han; methodology: **xiao Chen, Lang Wu, Guo Zhang, and **aoteng Fan; formal analysis and investigation: Tao Yan, Long Zhu, Yong**g Guan, Linjun Zhou, Tingting Hou, Xue Xue, **angju Li, Mingrong Wang, Haoran **ng, and **aofan **ong; writing original draft preparation: Xu Han; writing review and editing: Xu Han; funding acquisition: Lang Wu; resources: Lang Wu and Zaizhao Wang; supervision: Zaizhao Wang.
Corresponding author
Ethics declarations
Ethical approval
The current study does not include experiments involving living animals. All experimental procedures were approved by the Animal Welfare commissioner at the Department of Animal Science of the College of Animal Science and Technology, Northwest A&F University. All experiments were performed in accordance with relevant guidelines in China (Standards for the investigation of reservoir fishery resources, SL 167–2014).
Consent to participate
This research did not involve human participants.
Consent for publication
Our study did not use any kind of individual data such as videos and images. All authors have approved the final version for submission to your journal. All authors agreed to publication in the Journal of Environmental Science and Pollution Research.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Philippe Garrigues
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, X., Chen, J., Wu, L. et al. Species distribution modeling combined with environmental DNA analysis to explore distribution of invasive alien mosquitofish (Gambusia affinis) in China. Environ Sci Pollut Res 31, 25978–25990 (2024). https://doi.org/10.1007/s11356-024-32935-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-024-32935-5