Abstract
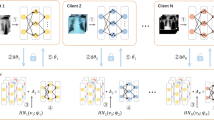
In tackling Tuberculosis (TB), a critical global health challenge, the integration of Foundation Models (FMs) into diagnostic processes represents a significant advance. FMs, with their extensive pre-training on diverse datasets, hold the promise of transforming TB diagnosis by leveraging their deep understanding and analytical capabilities. However, the application of these models in healthcare is complicated by the need to protect patient privacy, particularly when dealing with sensitive TB data from various medical centers. Our novel approach, FedARC, addresses this issue through personalized federated learning (PFL), enabling the use of private data without direct access. FedARC innovatively navigates data heterogeneity and privacy concerns by employing adaptive regularization and model-contrastive learning. This method not only aligns each center’s objective function with the global loss’s stationary point but also enhances model generalization across disparate data sources. Comprehensive evaluations on five publicly available chest X-ray image datasets demonstrate that foundation models profoundly influence outcomes, with our proposed method significantly surpassing contemporary methodologies in various scenarios.





Similar content being viewed by others
Availability of supporting Data
Not applicable.
References
WHO: Global tuberculosis report 2021. https://www.who.int/publications/digital/global-tuberculosis-report-2021, 1–57 (2021)
WHO: Global tuberculosis report 2022. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2022, 1–68 (2022)
Chauhan, A., Chauhan, D., Rout, C.: Role of gist and phog features in computer-aided diagnosis of tuberculosis without segmentation. PloS one 9(11), 112980 (2014)
Hwang, S., Kim, H.-E., Jeong, J., Kim, H.-J.: A novel approach for tuberculosis screening based on deep convolutional neural networks. In: Medical Imaging 2016: Computer-aided Diagnosis, vol. 9785, pp. 750–757 (2016). SPIE
Jaeger, S., Karargyris, A., Candemir, S., Folio, L., Siegelman, J., Callaghan, F., Xue, Z., Palaniappan, K., Singh, R.K., Antani, S., et al.: Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging 33(2), 233–245 (2013)
Candemir, S., Jaeger, S., Palaniappan, K., Musco, J.P., Singh, R.K., Xue, Z., Karargyris, A., Antani, S., Thoma, G., McDonald, C.J.: Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging. 33(2), 577–590 (2013)
Liu, Y., Wu, Y.-H., Ban, Y., Wang, H., Cheng, M.-M.: Rethinking computer-aided tuberculosis diagnosis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2646–2655 (2020)
Santosh, K., Allu, S., Rajaraman, S., Antani, S.: Advances in deep learning for tuberculosis screening using chest x-rays: The last 5 years review. J. Med. Syst. 46(11), 82 (2022)
McMahan, B., Moore, E., Ramage, D., Hampson, S., Arcas, B.A.: Communication-efficient learning of deep networks from decentralized data. In: Artificial Intelligence and Statistics, pp. 1273–1282 (2017). PMLR
Smith, V., Chiang, C.-K., Sanjabi, M., Talwalkar, A.S.: Federated multi-task learning. Adv. Neural Inf. Process. Syst. 30 (2017)
Arivazhagan, M.G., Aggarwal, V., Singh, A.K., Choudhary, S.: Federated learning with personalization layers. ar**v preprint ar**v:1912.00818 (2019)
Liang, P.P., Liu, T., Ziyin, L., Allen, N.B., Auerbach, R.P., Brent, D., Salakhutdinov, R., Morency, L.-P.: Think locally, act globally: Federated learning with local and global representations. ar**v preprint ar**v:2001.01523 (2020)
Li, X., Jiang, M., Zhang, X., Kamp, M., Dou, Q.: Fedbn: Federated learning on non-iid features via local batch normalization. ar**v preprint ar**v:2102.07623 (2021)
Collins, L., Hassani, H., Mokhtari, A., Shakkottai, S.: Exploiting shared representations for personalized federated learning. In: International Conference on Machine Learning, pp. 2089–2099 (2021). PMLR
Oh, J., Kim, S., Yun, S.-Y.: Fedbabu: Towards enhanced representation for federated image classification. ar**v preprint ar**v:2106.06042 (2021)
Chen, H.-Y., Chao, W.-L.: On bridging generic and personalized federated learning for image classification. ar**v preprint ar**v:2107.00778 (2021)
Wu, C., Wu, F., Lyu, L., Huang, Y., **e, X.: Communication-efficient federated learning via knowledge distillation. Nat. Commun. 13(1), 2032 (2022)
Xu, J., Tong, X., Huang, S.-L.: Personalized federated learning with feature alignment and classifier collaboration. In: The Eleventh International Conference on Learning Representations (2022)
Boykov, Y., Funka-Lea, G.: Graph cuts and efficient nd image segmentation. Int. J. Comput. Vis. 70(2) (2006)
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L.: Imagenet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp. 248–255 (2009). Ieee
Lopes, U., Valiati, J.F.: Pre-trained convolutional neural networks as feature extractors for tuberculosis detection. Comput. Biol. Med. 89, 135–143 (2017)
Rajaraman, S., Kim, I., Antani, S.K.: Detection and visualization of abnormality in chest radiographs using modality-specific convolutional neural network ensembles. PeerJ 8, 8693 (2020)
Liu, Y., Wu, Y.-H., Zhang, S.-C., Liu, L., Wu, M., Cheng, M.-M.: Revisiting computer-aided tuberculosis diagnosis. IEEE Trans. Pattern Anal. Mach. Intell. (2023)
Kairouz, P., McMahan, H.B., Avent, B., Bellet, A., Bennis, M., Bhagoji, A.N., Bonawitz, K., Charles, Z., Cormode, G., Cummings, R., et al.: Advances and open problems in federated learning. Found. Trends® Mach. Learn. 14(1–2), 1–210 (2021)
Li, X., Huang, K., Yang, W., Wang, S., Zhang, Z.: On the convergence of fedavg on non-iid data. ar**v preprint ar**v:1907.02189 (2019)
Zhu, Z., Hong, J., Zhou, J.: Data-free knowledge distillation for heterogeneous federated learning. In: International Conference on Machine Learning, pp. 12878–12889 (2021). PMLR
Jaeger, S., Candemir, S., Antani, S., Wáng, Y.-X.J., Lu, P.-X., Thoma, G.: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg. 4(6), 475 (2014)
Pan, C., Zhao, G., Fang, J., Qi, B., Liu, J., Fang, C., Zhang, D., Li, J., Yu, Y.: Computer-aided tuberculosis diagnosis with attribute reasoning assistance. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 623–633 (2022). Springer
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.-C.: Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Woo, S., Debnath, S., Hu, R., Chen, X., Liu, Z., Kweon, I.S., **e, S.: Convnext v2: Co-designing and scaling convnets with masked autoencoders. ar**v preprint ar**v:2301.00808 (2023)
Karimireddy, S.P., Kale, S., Mohri, M., Reddi, S., Stich, S., Suresh, A.T.: Scaffold: Stochastic controlled averaging for federated learning. In: International Conference on Machine Learning, pp. 5132–5143 (2020). PMLR
Li, Q., He, B., Song, D.: Model-contrastive federated learning. In: Proceedings of the IEEE/CVF Conference on computer vision and pattern recognition, pp. 10713–10722 (2021)
Funding
This work was supported in part by the National Key Reasearch and Development Program of China under Grants 2023YFC2705700, the National Natural Science Foundation of China under Grants 62225113, 62222112 and 62176186.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Special Issue on Advancing Recommendation Systems with Foundation Models
Guest Editors: Kai Zheng, Renhe Jiang, and Ryosuke Shibasaki
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, C., Luo, Y., Xu, Y. et al. Foundation models matter: federated learning for multi-center tuberculosis diagnosis via adaptive regularization and model-contrastive learning. World Wide Web 27, 30 (2024). https://doi.org/10.1007/s11280-024-01266-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11280-024-01266-3