Abstract
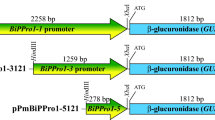
NAM/ATAF/CUC (NAC) family genes comprise one of the largest families of transcription factors in plant genomes and are widely expressed in develo** woody tissues. In the present study, we constructed plant transformation vectors using the β-glucuronidase (GUS) reporter gene system and detected the promoter expression patterns derived from the PtNAC068 and PtNAC154 genes of Chinese white poplar (Populus tomentosa Carr.) in transgenic poplars (Populus alba × Populus glandulosa). The results showed that the GUS expression driven by PtNAC068 and PtNAC154 promoters may be more complex in poplar than they are in Arabidopsis. Histochemical GUS assays showed that GUS activity driven by PtNAC068 promoter was mainly in vascular tissues of stems, leaves, petioles, and roots, while that driven by PtNAC154 promoter was confined to the develo** secondary xylem of stems and veins of leaves. The transcript level of both PtNAC068 and PtNAC154 in successive internodes below the apex was found to be much higher in IN5-10 compared to that in IN2-4 as measured by real-time RT-PCR, suggesting that PtNAC068 and PtNAC154 upregulation is related to secondary growth in poplar. GUS expression in internodes 3–8 of ProNAC068::GUS transgenic plants was 30-fold higher than that in ProNAC154::GUS transgenic plants. The differences in the expression pattern and transcript level of mRNA accumulation indicate that PtNAC068 and PtNAC154 may be involved in two distinct aspects of vascular tissue development.




Similar content being viewed by others
Abbreviations
- aa:
-
Amino acid
- CTAB:
-
Cetyl trimethylammonium bromide
- GUS:
-
β-Glucuronidase
- IN:
-
Internode
- NAC:
-
NAM/ATAF/CUC
- NPTII:
-
Neomycin phosphotransferase II
- RT-qPCR:
-
Real-time quantitative PCR
- X-gluc:
-
5-Bromo-4-chloro-3-indoxyl-beta-d-glucuronic acid
References
Candela H, Martinez-Laborda A, Micol JL (1999) Venation pattern formation in Arabidopsis thaliana vegetative leaves. Dev Biol 205:205–216
Demura T, Fukuda H (2007) Transcriptional regulation in wood formation. Trends Plant Sci 12:64–70
Dengler N, Kang J (2001) Vascular patterning and leaf shape. Curr Opin Plant Biol 4:50–56
Dharmawardhana P, Brunner AM, Strauss SH (2010) Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa. BMC Genomics 11:150–168
Du J, Groover A (2010) Transcriptional regulation of secondary growth and wood formation. J Integr Plant Biol 52:17–27
Du J, **e HL, Zhang DQ, He XQ, Wang MJ, Li YZ, Cui KM, Lu MZ (2006) Regeneration of the secondary vascular system in poplar as a novel system to investigate gene expression by a proteomic approach. Proteomics 6:881–895
Duval M, Hsieh TF, Kim SY, Thomas TL (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248
Ernst HA, Olsen AN, Larsen S, Lo Leggio L (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5:297–303
Fan J, Gao X, Yang YW, Deng W, Li ZG (2007) Molecular cloning and characterization of a NAC-like gene in “navel” orange fruit response to postharvest stresses. Plant Mol Biol Rep 25:145–153
Fang Y, You J, **e K, **e W, **ong L (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol Genet Genom 280:547–563
Grant EH, Fu**o T, Beers EP, Brunner AM (2010) Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232:337–352
Guo YF, Gan SS (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612
Hawkins S, Samaj J, Lauvergeat V, Boudet A, Grima-Pettenati J (1997) Cinnamyl alcohol dehydrogenase: identification of new sites of promoter activity in transgenic poplar. Plant Physiol 113:321–325
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27:297–300
Hu R, Qi G, Kong Y, Kong D, Gao Q, Zhou G (2010) Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol 10:145–167
Jensen MK, Kjaersgaard T, Petersen K, Skriver K (2010) NAC genes: time-specific regulators of hormonal signaling in Arabidopsis. Plant Signal Behav 5:907–910
Ko JH, Beers EP, Han KH (2006) Global comparative transcriptome analysis identifies gene network regulating secondary xylem development in Arabidopsis thaliana. Mol Genet Genom 276:517–531
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50:1035–1048
Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19:1855–1860
Liu YZ, Baig MNR, Fan R, Ye JL, Cao YC, Deng XX (2009) Identification and expression pattern of a novel NAM, ATAF, and CUC-like gene from Citrus sinensis Osbeck. Plant Mol Biol Rep 27:292–297
Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19:270–280
Mitsuda N, Ohme-Takagi M (2008) NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J 56:768–778
Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci 10:79–87
Pfaffl M (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007
Phillips MA, D’Auria JC, Luck K, Gershenzon J (2009) Evaluation of candidate reference genes for real-time quantitative PCR of plant samples using purified cDNA as template. Plant Mol Biol Rep 27:407–416
Pinheiro GL, Marques CS, Costa MD, Reis PA, Alves MS, Carvalho CM, Fietto LG, Fontes EP (2009) Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 444:10–23
Plomion C, Leprovost G, Stokes A (2001) Wood formation in trees. Plant Physiol 127:1513–1523
Rushton PJ, Bokowiec MT, Han S, Zhang H, Brannock JF, Chen X, Laudeman TW, Timko MP (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol 147:280–295
Sambrook J, Russell D (2001) Molecular cloning, a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, New York
Shen H, Yin Y, Chen F, Xu Y, Dixon R (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioenerg Res 2:217–232
Taoka K, Yanagimoto Y, Daimon Y, Hibara K, Aida M, Tasaka M (2004) The NAC domain mediates functional specificity of CUP-SHAPED COTYLEDON proteins. Plant J 40:462–473
Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Wang M, Qi X, Zhao S, Zhang S, Lu MZ (2009) Dynamic changes in transcripts during regeneration of the secondary vascular system in Populus tomentosa Carr. revealed by cDNA microarrays. BMC Genomics 10:215–222
**e Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Yamaguchi M, Demura T (2010) Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol 27:237–242
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) Vascular-related NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55:652–664
Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol 138:803–818
Zhong R, Lee C, Ye ZH (2009) Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 152:1044–1055
Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225:1603–1611
Zhong R, Ye ZH (2010) The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis. Plant Signal Behav 5:469–472
Zhou Z, Wang MJ, Zhao ST, Hu JJ, Lu MZ (2009) Changes in freezing tolerance in hybrid poplar caused by up- and down-regulation of PtFAD2 gene expression. Transgenic Res 19:647–654
Acknowledgement
The work presented here was supported by the Natural National Science Foundation of China (31030018).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Han, X., He, G., Zhao, S. et al. Expression Analysis of Two NAC Transcription Factors PtNAC068 and PtNAC154 from Poplar. Plant Mol Biol Rep 30, 370–378 (2012). https://doi.org/10.1007/s11105-011-0350-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0350-1