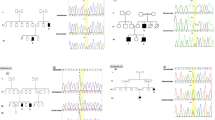
Abstract
Studies of X-linked pedigrees were the first to identify genes implicated in intellectual disability (ID) and autism spectrum disorder (ASD). However, some pedigrees present a huge clinical variability between the affected members. This intrafamilial heterogeneity may be due to cooccurrence of two disorders. In the present study, we describe a multiplex X-linked pedigree in which three siblings have ID, ASD and dysmorphic features but with variable severity. Through Fragile X syndrome test, we identified the full FMR1 mutation in only two males. Whole exome sequencing allowed us to identify a novel hemizygous variant (p.Gln2080_Gln2083del) in MED12 gene in two males. So, the first patient has FXS, the second has both FMR1 and MED12 mutations while the third has only the MED12 variant. MED12 mutations are implicated in several forms of X-linked ID. Family segregation and genotype–phenotype–correlation were in favor of a cooccurrence of two forms of X-linked ID. Our work provides further evidence of the involvement of MED12 in XLID. Moreover, through these results, it is noteworthy to raise awareness that intrafamilial heterogeneity in FXS multiplex families could result from the cooccurrence of multiple clinical entities involving at least two separate genetic loci. This should be taken into consideration for genetic testing and counselling in patients/families with atypical disease symptoms.



Similar content being viewed by others
References
Lubs HA, Stevenson RE, Schwartz CE (2012) Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet 90:579–590. https://doi.org/10.1016/j.ajhg.2012.02.018
Crawford DC, Meadows KL, Newman JL et al (2002) Prevalence of the fragile X syndrome in African–Americans. Am J Med Genet 110:226–233. https://doi.org/10.1002/ajmg.10427
Kaufmann WE, Reiss AL (1999) Molecular and cellular genetics of fragile X syndrome. Am J Med Genet 88:11–24
Pieretti M, Zhang F, Fu Y-H et al (1991) Absence of expression of the FMR-1 gene in fragile X syndrome. Cell 66:817–822
Hessl D, Dyer-Friedman J, Glaser B et al (2001) The influence of environmental and genetic factors on behavior problems and autistic symptoms in boys and girls with fragile X syndrome. Pediatrics 108:e88–e88
McDuffie A, Thurman AJ, Hagerman RJ, Abbeduto L (2015) Symptoms of autism in males with fragile x syndrome: a comparison to nonsyndromic ASD using current ADI-R scores. J Autism Dev Disord 45:1925–1937. https://doi.org/10.1007/s10803-013-2013-6
American Psychiatric Association (2013) Diagnostic and statistical manual of mental disorders (DSM-5®). American Psychiatric Pub
Clifford S, Dissanayake C, Bui QM et al (2007) Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord 37:738–747
Rogers SJ, Wehner EA, Hagerman R (2001) The behavioral phenotype in fragile X: symptoms of autism in very young children with fragile X syndrome, idiopathic autism, and other developmental disorders. J Dev Behav Pediatr 22:409–417
Hatton DD, Hooper SR, Bailey DB et al (2002) Problem behavior in boys with fragile X syndrome. Am J Med Genet 108:105–116
Hatton DD, Sideris J, Skinner M et al (2006) Autistic behavior in children with fragile X syndrome: prevalence, stability, and the impact of FMRP. Am J Med Genet A 140A:1804–1813. https://doi.org/10.1002/ajmg.a.31286
Kau AS, Tierney E, Bukelis I et al (2004) Social behavior profile in young males with fragile X syndrome: characteristics and specificity. Am J Med Genet A 126:9–17
Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24:659–685
Lord C, Rutter M, DiLavore P, et al (2012) Autism Diagnostic Observation Schedule Second Edition (ADOS-2) Manual (Part 1): Modules 1–4. Torrance CA West Psychol Serv
Carter AS, Volkmar FR, Sparrow SS et al (1998) The Vineland adaptive behavior scales: supplementary norms for individuals with autism. J Autism Dev Disord 28:287–302
Perron-Borelli M (2000) EDEI R: Echelles Différentielles d’Efficiences Intellectuelles: forme révisée: Manuel. Editions EAP
Fu Y-H, Kuhl DP, Pizzuti A et al (1991) Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell 67:1047–1058
Panagopoulos I, Lassen C, Kristoffersson U, Åman P (1999) A methylation PCR approach for detection of fragile X syndrome. Hum Mutat 14:71–79
Van der Auwera GA, Carneiro MO, Hartl C et al (2013) From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma 43:11
Wang W, Lufkin T (2000) The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the develo** hypothalamus. Dev Biol 227:432–449
Wang X, Yang N, Uno E et al (2006) A subunit of the mediator complex regulates vertebrate neuronal development. Proc Natl Acad Sci 103:17284–17289
Spikol ED, Glasgow E (2018) Separate roles for Med12 and Wnt signaling in regulation of oxytocin expression. Biol Open 7:bio031229. https://doi.org/10.1242/bio.031229
Risheg H, Graham JM Jr, Clark RD et al (2007) A recurrent mutation in MED12 leading to R961 W causes Opitz-Kaveggia syndrome. Nat Genet 39:451–453
Schwartz CE, Tarpey PS, Lubs HA et al (2007) The original Lujan syndrome family has a novel missense mutation (p N1007S) in the MED12 gene. J Med Genet 44(7):472–477
Vulto-van Silfhout AT, de Vries BB, van Bon BW et al (2013) Mutations in MED12 cause X-linked Ohdo syndrome. Am J Hum Genet 92:401–406
Graham JM, Schwartz CE (2013) MED12 related disorders. Am J Med Genet A. https://doi.org/10.1002/ajmg.a.36183
Margolis RL, Abraham MR, Gatchell SB et al (1997) cDNAs with long CAG trinucleotide repeats from human brain. Hum Genet 100:114–122
Butland SL, Devon RS, Huang Y et al (2007) CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics 22(8):126. https://doi.org/10.1186/1471-2164-8-126
DeLisi LE, Smith AB, Razi K et al (2000) Investigation of a candidate gene for schizophrenia on Xq13 previously associated with mental retardation and hypothyroidism. Am J Med Genet 96:398–403
Philibert RA (2006) A meta-analysis of the association of the HOPA12 bp polymorphism and schizophrenia. Psychiatr Genet 16:73–76
Philibert RA, Bohle P, Secrest D et al (2007) The association of the HOPA12 bp polymorphism with schizophrenia in the NIMH genetics initiative for schizophrenia sample. Am J Med Genet B Neuropsychiatr Genet 144:743–747
Spinks R, Sandhu HK, Andreasen NC, Philibert RA (2004) Association of the HOPA12 bp allele with a large X-chromosome haplotype and positive symptom schizophrenia. Am J Med Genet B Neuropsychiatr Genet 127:20–27
Beyer KS, Klauck SM, Benner A et al (2002) Association studies of the HOPA dodecamer duplication variant in different subtypes of autism. Am J Med Genet 114:110–115
Philibert RA, Winfield SL, Sandhu HK et al (2000) The structure and expression of the human neuroligin-3 gene. Gene 246:303–310
Graham JM Jr, Visootsak J, Dykens E et al (2008) Behavior of 10 patients with FG syndrome (Opitz–Kaveggia syndrome) and the p. R961W mutation in the MED12 gene. Am J Med Genet A 146:3011–3017
Fieremans N, Van Esch H, Holvoet M et al (2016) Identification of intellectual disability genes in female patients with a skewed X-inactivation pattern. Hum Mutat 37:804–811
Langley KG, Brown J, Gerber RJ et al (2015) Beyond Ohdo syndrome: a familial missense mutation broadens the MED12 spectrum. Am J Med Genet A 167:3180–3185. https://doi.org/10.1002/ajmg.a.37354
Kraus DM, Elliott GS, Chute H et al (2006) CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol 176:4419–4430
Håvik B, Le Hellard S, Rietschel M et al (2011) The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry 70:35–42
Liu W, Liu F, Xu X, Bai Y (2017) Replicated association between the European GWAS locus rs10503253 at CSMD1 and schizophrenia in Asian population. Neurosci Lett 647:122–128
Xu W, Cohen-Woods S, Chen Q et al (2014) Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med Genet 15:2
Athanasiu L, Giddaluru S, Fernandes C et al (2017) A genetic association study of CSMD1 and CSMD2 with cognitive function. Brain Behav Immun 61:209–216
Liu X, Shimada T, Otowa T et al (2016) Genome-wide association study of autism spectrum disorder in the east Asian populations. Autism Res 9:340–349
Cukier HN, Dueker ND, Slifer SH et al (2014) Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism 5:1
Krumm N, Turner TN, Baker C et al (2015) Excess of rare, inherited truncating mutations in autism. Nat Genet 47:582–588. https://doi.org/10.1038/ng.3303
Mahmoudi E, Cairns MJ (2016) MiR-137: an important player in neural development and neoplastic transformation. Mol Psychiatry 22:44–55
Donohoe G, Walters J, Hargreaves A et al (2013) Neuropsychological effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. Genes Brain Behav 12:203–209
Acknowledgements
We are grateful to the family members for their collaboration. This work was supported by the Tunisian Ministry of Public Health and by the Ministry of Higher Education and Scientific Research (LR16IPT05).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
All procedures performed in this study were approved by the Institute Pasteur of Tunis ethics committee (Reference: 2016/17/I/LR11IPT05) and were in accordance with the Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
The participants to the study provided their written informed consent. Informed consent was obtained from the mother, the legal guardian of the minors involved in the study for their participation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lahbib, S., Trabelsi, M., Dallali, H. et al. Novel MED12 variant in a multiplex Fragile X syndrome family: dual molecular etiology of two X-linked intellectual disabilities with autism in the same family. Mol Biol Rep 46, 4185–4193 (2019). https://doi.org/10.1007/s11033-019-04869-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-04869-6