Abstract
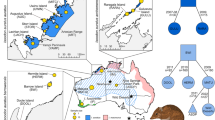
Understanding population genetic structure and levels of genetic variation is critical for the conservation and management of imperiled populations, especially when reintroductions are planned. We used restriction-site associated DNA (RAD) sequencing to study the genetic diversity and evolutionary relationships of the endangered Amargosa vole and other closely related desert-dwelling California voles. Specifically, we sought to determine how Amargosa voles are related to other California voles, how genetic variation is partitioned among subpopulations in wild Amargosa voles, and how much genetic variation is captured within a captive insurance colony of Amargosa voles. Our multilocus nuclear dataset provides strong evidence that Amargosa voles are part of a northern clade of California voles. Amargosa voles have highly reduced genetic variation relative to other California voles, but do exhibit some sub-structure among sampled marshes. Captive Amargosa voles capture approximately half of the total genetic variation present in the wild Amargosa vole populations. We discuss the management implications of our findings in light of reintroductions planned for Amargosa voles. Our study highlights the utility of reduced representation genomic approaches, like RADseq, to resolve relationships among small populations that are difficult to study with traditional markers due to low genetic variation and few individuals left in the wild.







Similar content being viewed by others
References
Ali OA, O’Rourke SM, Amish SJ et al (2016) Rad capture (rapture): flexible and efficient sequence-based genoty**. Genetics 202:389–400. doi: 10.1534/genetics.115.183665
Barley AJ, Monnahan PJ, Thomson RC et al (2015) Sun skink landscape genomics: assessing the roles of micro-evolutionary processes in sha** genetic and phenotypic diversity across a heterogeneous and fragmented landscape. Mol Ecol 24:1696–1712. doi: 10.1111/mec.13151
Bowen BS (1982) Temporal dynamics of microgeographic structure of genetic variation in Microtus californicus. J Mammal 63:625–638
Bradburd GS, Ralph PL, Coop GM (2016) A spatial framework for understanding population structure and admixture. PLoS Genet 12:1–38. doi: 10.1371/journal.pgen.1005703
Catchen J, Hohenlohe PA, Bassham S et al (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. doi: 10.1111/mec.12354
Conroy CJ, Neuwald JL (2008) Phylogeographic study of the California vole, Microtus californicus. J Mammal 89:755–767. doi: 10.1644/07-MAMM-A-189R1.1
Conroy CJ, Patton JL, Lim MCW et al (2016) Following the rivers: historical reconstruction of California voles Microtus californicus (Rodentia: Cricetidae) in the deserts of eastern California. Biol J Linn Soc 119:80–98. doi: 10.1111/bij.12808
Di Candia MR, Routman EJ (2007) Cytonuclear discordance across a leopard frog contact zone. Mol Phylogenet Evol 45:564–575. doi: 10.1016/j.ympev.2007.06.014
Do C, Waples RS, Peel D et al (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14:209–214. doi: 10.1111/1755-0998.12157
Docker MF, Heath DD (2003) Genetic comparison between sympatric anadromous steelhead and freshwater resident rainbow trout in British Columbia, Canada. Conserv Genet 4:227–231. doi: 10.1023/A:1023355114612
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Foley J (2016) Annual performance report: captive breeding of a model species and reconstructing habitat ahead of translocation of the critically endangered Amargosa vole. Presented to BLM and USFWS
Fumagalli M, Vieira FG, Korneliussen TS et al (2013) Quantifying population genetic differentiation from next-generation sequencing data. Genetics 195:979–992. doi: 10.1534/genetics.113.154740
Funk WC, Lovich RE, Hohenlohe PA et al (2016) Adaptive divergence despite strong genetic drift: genomic analysis of the evolutionary mechanisms causing genetic differentiation in the island fox (Urocyon littoralis). Mol Ecol 25:2176–2194. doi: 10.1111/mec.13605
Gill AE (1984) Partial reproductive isolation of subspecies of the California vole, Microtus californicus. Genetica 52–53:105–117
Gottscho AD, Wood DA, Vandergast AG et al (2017) Lineage diversification of fringe-toed lizards (Phrynosomatidae: Uma notata complex) in the Colorado Desert: delimiting species in the presence of gene flow. Mol Phylogenet Evol 106:103–117. doi: 10.1016/j.ympev.2016.09.008
Hall ER (1981) The mammals of North America. John Wiley & Sons, New York
Hartl DL, Clark AG (1997) Principles of population genetics, 3rd edn. Sinauer Associates, Sunderland, 542 pp
Hill W (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res Camb 38:209–216. doi: 10.1017/S0016672300020553
Jezkova T, Riddle BR, Card DC et al (2015) Genetic consequences of postglacial range expansion in two codistributed rodents (genus Dipodomys) depend on ecology and genetic locus. Mol Ecol 24:83–97. doi: 10.1111/mec.13012
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632. doi: 10.1038/hdy.2010.95
Kardos M, Luikart G, Allendorf FW (2015) Measuring individual inbreeding in the age of genomics: marker-based measures are better than pedigrees. Heredity 115:63–72. doi: 10.1038/hdy.2015.17
Kellog R (1918) A revision of the Microtus californicus group of meadow mice. Univ Calif Publ Zool 21:1–42
Klinger R (2015) Estimation of the total population size of the Amargosa vole. Report to United States Geological Survey, Bishop
Klinger R, Cleaver M, Anderson S, Maier P, Clark J (2013) Short-term population dynamics, demography, and habitat selection by the Amargosa vole. Report to the BLM. United States Geological Survey, Western Ecological Research Station, Bishop
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. doi: 10.1111/1755-0998.12387
Korneliussen TS, Moltke I (2015) NgsRelate: a software tool for estimating pairwise relatedness from next-generation sequencing data. Bioinformatics 31:4009–4011. doi: 10.1093/bioinformatics/btv509
Korneliussen TS, Moltke I, Albrechtsen A, Nielsen R (2013) Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinform 14:289. doi: 10.1186/1471-2105-14-289
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinform 15:356. doi: 10.1186/s12859-014-0356-4
Krebs CJ (1966) Demographic changes in fluctuating populations of Microtus californicus. Ecol Monogr 36:239–273. doi: 10.2307/1942418
La Haye MJJ, Reiners TE, Raedts R et al (2017) Genetic monitoring to evaluate reintroduction attempts of a highly endangered rodent. Conserv Genet. doi: 10.1007/s10592-017-0940-z
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Lindell J, Méndez-De La Cruz FR, Murphy RW (2008) Deep biogeographical history and cytonuclear discordance in the black-tailed brush lizard (Urosaurus nigricaudus) of Baja California. Biol J Linn Soc 94:89–104. doi: 10.1111/j.1095-8312.2008.00976.x
Madsen T, Shine R, Olsson M, Wittzell H (1999) Conservation biology: restoration of an inbred adder population. Nature 402:34–35. doi: 10.1038/46941
Magoč T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507°
McKenna A, Hanna M, Banks E et al (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110
Miller W, Wright SJ, Zhang Y et al (2010) Optimization methods for selecting founder individuals for captive breeding or reintroduction of endangered species. Pac Symp Biocomput 53:43–53. doi: 9789814295291_0006
Murdoch D, Chow ED (2013) ellipse: Functions for drawing ellipses and ellipse-like confidence regions. R package version 0.3-8. https://CRAN.R-project.org/package=ellipse
Neuwald JL (2010) Population isolation exacerbates conservation genetic concerns in the endangered Amargosa vole, Microtus californicus scirpensis. Biol Conserv 143:2028–2038. doi: 10.1016/j.biocon.2010.05.007
Norton JE, Ashley MV (2004) Genetic variability and population differentiation in captive Baird’s tapirs (Tapirus bairdii). Zoo Biol 23:521–531. doi: 10.1002/zoo.20031
Pagès M, Bazin E, Galan M et al (2013) Cytonuclear discordance among Southeast Asian black rats (Rattus rattus complex). Mol Ecol 22:1019–1034. doi: 10.1111/mec.12149
Petkova D, Novembre J, Stephens M (2016) Visualizing spatial population structure with estimated effective migration surfaces. Nat Genet 48:94–100. doi: 10.1101/011809
Prunier JG, Kaufmann B, Fenet S et al (2013) Optimizing the trade-off between spatial and genetic sampling efforts in patchy populations: towards a better assessment of functional connectivity using an individual-based sampling scheme. Mol Ecol 22:5516–5530. doi: 10.1111/mec.12499
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. doi: 10.1111/1755-0998.12512
R Core Team (2016). R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rick J (2015) Genetic diversity, structure, and hybridization in a harvested gray wolf (Canis lupus) population in Minnesota. MS Thesis, University of Minnesota
Singhal S, Moritz C (2012) Testing hypotheses for genealogical discordance in a rainforest lizard. Mol Ecol 21:5059–5072. doi: 10.1111/j.1365-294X.2012.05747.x
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics 195:693–702. doi: 10.1534/genetics.113.154138
Tallmon DA, Luikart G, Waples RS (2004) The alluring simplicity and complex reality of genetic rescue. Trends Ecol Evol 19:489–496. doi: 10.1016/j.tree.2004.07.003
Taylor ZS, Hoffman SMG (2012) Microsatellite genetic structure and cytonuclear discordance in naturally fragmented populations of deer mice (Peromyscus maniculatus). J Hered 103:71–79. doi: 10.1093/jhered/esr100
Teng H, Zhang Y, Shi C et al (2016) Whole-genome sequencing reveals genetic variation in the Asian House Rat. G3 6:1969–1977. doi: 10.1534/g3.116.029504
Tordoff HB, Redig PT (2001) Role of genetic background in the success of reintroduced peregrine falcons. Conserv Biol 15:528–532. doi: 10.1046/j.1523-1739.2001.015002528.x
United States Fish and Wildlife Service (2009) Amargosa vole Microtus californicus scirpensis 5-year review: summary and evaluation. United States Fish and Wildlife Service, Ventura, p. 29
Wang J (2017) The computer program structure for assigning individuals to populations: easy to use but easier to misuse. Mol Ecol. doi: 10.1111/1755-0998.12650
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262. doi: 10.1111/j.1752-4571.2009.00104.x
Weir BS, Anderson AD, Hepler AB (2006) Genetic relatedness analysis: modern data and new challenges. Nat Rev Genet 7:771–780. doi: 10.1038/nrg1960
Wright S (1943) Isolation by distance. Genetics 28:114–138
Yang D-S, Kenagy GJ (2009) Nuclear and mitochondrial DNA reveal contrasting evolutionary processes in populations of deer mice (Peromyscus maniculatus). Mol Ecol 18:5115–5125. doi: 10.1111/j.1365-294X.2009.04399.x
Acknowledgements
Funding was provided by the Bureau of Land Management (SPO Number 201223906). We thank Andrew DeWoody, and the DeWoody lab at Purdue University for access to a draft Amargosa vole genome. We thank the Museum of Vertebrate Zoology (UC Berkeley) for access to tissues of California voles, and Jim Patton for capturing and giving insight on Microtus generally. We thank two anonymous reviewers and Rosemary Gillespie for their helpful feedback on this manuscript. This work was approved by the UC Davis IACUC (Permit Numbers 19741 and 18179).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10592_2017_1010_MOESM2_ESM.eps
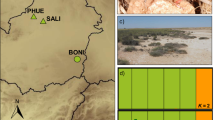
Supplementary material 2—NGSadmix analyses for other K values for California voles, including Amargosa voles. Admixture is shown on the y-axis and each major grou** of voles, from the northern, southern or Amargosa clade, is shown on the x-axis. Dashed black lines separate geographic sampling locations for the NGSadmix plots, while dashed white lines separate the grou**s of each K. (EPS 1464 KB)
10592_2017_1010_MOESM3_ESM.eps
Supplementary material 3—NGSadmix analyses for other K values for wild Amargosa voles. Admixture is on the y-axis and marshes sampled is on the x-axis. Dashed white lines separate marshes sampled in this study. (EPS 1377 KB)
10592_2017_1010_MOESM4_ESM.eps
Supplementary material 4—NGSadmix analyses for other K values for wild and captive Amargosa voles. Admixture is on the y-axis and marshes or captive colony generation sampled is on the x-axis. Dashed white lines separate marshes or captive colony generation sampled in this study. (EPS 1538 KB)
Rights and permissions
About this article
Cite this article
Krohn, A.R., Conroy, C.J., Pesapane, R. et al. Conservation genomics of desert dwelling California voles (Microtus californicus) and implications for management of endangered Amargosa voles (Microtus californicus scirpensis). Conserv Genet 19, 383–395 (2018). https://doi.org/10.1007/s10592-017-1010-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-017-1010-2