Abstract
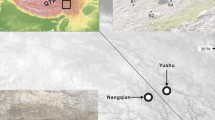
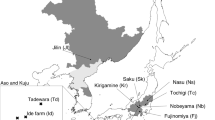
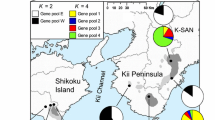
Tricyrtis ishiiana is a relic endemic plant taxon of the Convallariaceae that inhabits two nearby gorges in Kanagawa Prefecture, Japan. The distribution range and number of populations are thought to have been reduced to the present refugial populations during the Quaternary climatic oscillations. Because of its showy flowers, this plant has faced illegal removal from its natural habitats for horticultural use and has been designated a critically endangered species (class IA). In this study, we analyzed the genetic structure of the relict populations of T. ishiiana in order to contribute to the conservation strategies of the prefectural government. Our analyses of nine nuclear microsatellite loci detected high genetic diversity (HE = 0.704 and HO = 0.541) for the two populations. The two populations were slightly differentiated (RST = 0.032), accompanied by faint substructure across the populations (K = 3). In addition, each population exhibited spatial genetic structuring. The relatively low inbreeding coefficient for both populations together (FIS = 0.233) and each population separately (FIS = 0.217–0.246) may be attributable to crossing among descendants within a population along with occasional gene flow between the populations. These results suggested that the extant populations have not experienced a severe bottleneck. The two extant populations were genetically differentiated at a very low level, accompanied by occasional pollen flow via pollinators and/or seed dispersal by gravity in the mountainous environment. Occasional gene exchange between the populations has allowed T. ishiiana to harbor high genetic diversity despite being a relic plant confined to two small refugial populations.





Similar content being viewed by others
References
Cornuet JM, Luikart G (1996) Description and evaluation of two tests for detecting recent bottlenecks. Genetics 144:2001–2014
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from plant tissue. Focus 12:13–15
Edwards AL, Wyatt R (1994) Population genetics of the rare Asclepias texana and its widespread sister species, A. perennis. Syst Bot 19:291–307
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor Appl Genet 92:832–839
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Godt MJW, Walker J, Hamrick JL (1997) Genetic diversity in the endangered lily Harperocallis flava and a close relative, Tofieldia racemosa. Conser Biol 11:361–366
Goudet J (1995) Fstat version 1.2: a computer program to calculate F statistics. J Hered 86:485–486
Goudet J, Raymond M, De-Meeus T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Hardy OJ, Vekemans X (2002) SPAGEDI: a versatile computer program to analyze spatial genetic structure at the individual or population level. Mol Ecol Notes 2:618–620
Ikeda H, Senni K, Fujii N, Setoguchi H (2006) Refugia of Potentilla matsumurae (Rosaceae) located at high mountains in the Japanese archipelago. Mol Ecol 15:3731–3740
Japan Society of Plant Taxonomists (1993) Red Data Book: Japanese endangered plants. Nousonbunka-sha, Tokyo (in Japanese)
Linhart YB, Permoli AC (1993) Genetic variation in Altes acaulis and its relative, the narrow endemic A. humilis (Apiaceae). Am J Bot 80:598–605
Luikart G, Allendorf FW, Cornuet JM, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89:238–247
Lumaret R, Mir C, Michaud H, Raynal V (2002) Phylogeograpical variation of chloroplast DNA in holm oak (Quercus ilex L.). Mol Ecol 11:2327–2336
Maki Y, Morita H, Oiki S, Takahashi H (1999) The effect of geographic range and dichogamy on genetic variability and population genetic structure in Tricyrtis section Flavae (Liliaceae). Am J Bot 86:287–292
Maruyama T, Fuerst PA (1985) Population bottlenecks and non equilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–678
Ministry of the Environment of Japan (2000) Threatened wildlife of Japan, Red Data Book, vol 8, 2nd ed (vascular plants). Japan Wildlife Research Center, Tokyo (in Japanese)
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Palmé AE, Vendramin GG (2002) Chloroplast DNA variation, postglacial recolonization and hybridization in hazel, Corylus avellana. Mol Ecol 11:1769–1779
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petit RJ, Brewer S, Bordács S, Burg K, Cheddadi R, Coart E, Cottrell J, Csaikl UM, van Dam B, Deans JD, Espinel S, Fineschi S, Finkeldey R, Glaz I, Goicoechea PG, Jensen JS, König AO, Lowe AJ, Madsen SF, Mátyás G, Munro RC, Popescu F, Slade D, Tabbener H, de Vries SGM, Ziegenhagen B, de Beaulieu JL, Kremer A (2002) Identification of refugia and post-glacial colonization routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For Ecol Manag 156:49–74
Petit RJ, Aguinagalde I, de Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Muller-Starck GM, Demesure-Musch B, Palme A, Martin JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Piry S, Luikart G, Cornuet JM (1999) Bottleneck: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Purdy BG, Bayer RJ (1995a) Allozyme variation in the Athabasca sand dune endemic, Salix silicicola, and the closely related widespread species, S. alaxensis. Syst Bot 20:179–190
Purdy BG, Bayer RJ (1995b) Genetic diversity in the tetraploid sand dune endemic Deshampia mackenzieana and its widespread progenitor D. cespitosa (Poaceae). Am J Bot 82:121–130
Purdy BG, Bayer RJ (1996) Genetic variation in populations of the endemic Achillea millefolium ssp. megacephala from the Athabasca sand dunes and the widespread ssp. lanulosa in western North America. Can J Bot 74:1138–1146
Qiu YX, Sun Y, Zhang XP, Lee J, Fu CX, Comes HP (2009) Molecular phylogeography of East Asian Kirengeshoma (Hydrangeaceae) in relation to quaternary climate change and landbridge configurations. New Phytol 183:480–495
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evol Int J Org Evol 43:258–275
Rousset F (1996) Equilibrium values of measures of population subdivision for stepwise mutation processes. Genetics 142:1357–1362
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Schönswetter P, Stehlik I, Holderegger R, Tribsch A (2005) Molecular evidence for glacial refugia of mountain plants in the Europe Alps. Mol Ecol 14:3547–3555
Setoguchi H, Ohba H (1995) Phylogenetic relationships in Crossostylis inferred from restriction site variation of chloroplast DNA. J Plant Res 108:87–92
Setoguchi H, Mitsui Y, Ikeda H, Nomura N, Tamura A (2009) Development and characterization of microsatellite loci in the endangered Tricyrtis ishiiana (Convallariaceae), a local endemic plant in Japan. Conserv Genet 10:705–707
Sherman-Broyles SL, Gibson JP, Hamrick JL, Bucher MA, Gibson MJ (1992) Compariosn of allozyme diversity among rare and widespread Rhus species. Syst Bot 17:551–559
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequency. Genetics 139:457–462
Takahashi H (1993) Floral biology of Tricyrtis macranthopsis Masamune and T. ishiiana (Kitagawa et T. Koyama) Ohwi et Okuyama (Liliaceae). Acta Phytotaxonomica et Geobotanica 44:141–150
Takahashi Y, Takahashi H, Maki Y Comparison of genetic variation and differentiation using microsatellite markers among three rare threatened and one widespread toad lily species of Tricyrtis section Flavae (Convallariaceae) in Japan. Plant Species Biol (in press)
Watterson GA (1978) The homozygosity test of neutrality. Genetics 88:405–417
Watterson GA (1986) The homozygosity test after a change in population size. Genetics 112:899–907
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
We are grateful to Ms. Akiko Sakai (Kanagawa Prefecture Natural Environment Conservation Center), and Asuka Noda, Kanako Sugahara and Tomomi Dan (Kyoto University) for their cooperation in collecting materials and extracting DNA, respectively. Prof. Masayuki Maki (Tohoku University) kindly showed us his unpublished data (currently in press). This study was supported by a grant for Conservation Genetics for Endangered Plants from Kanagawa Prefecture Natural Environment Conservation Center.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Setoguchi, H., Mitsui, Y., Ikeda, H. et al. Genetic structure of the critically endangered plant Tricyrtis ishiiana (Convallariaceae) in relict populations of Japan. Conserv Genet 12, 491–501 (2011). https://doi.org/10.1007/s10592-010-0156-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0156-y