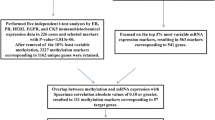
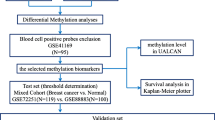
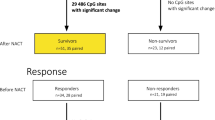
Abstract
Breast cancer patients with similar clinical stage may experience different disease outcomes. Aberrant DNA methylation of primary breast tumors can have impact on the clinical outcome. This study aimed to assess clinical utility of tumor-specific methylated sequences (MINT17, 31) and tumor-related gene (RARβ2) methylation classification in primary breast tumors. Absolute quantitative assessment of methylated alleles (AQAMA) was used to determine the methylation index (MI) of MINT17, MINT31, and RARß2 in 242 primary tumors of early stage breast cancer patients. Patients were classified into three methylation groups: meth-N, with normal methylation levels of all biomarkers; meth-L, with one biomarker hypermethylation; and meth-H, with hypermethylation of >1 biomarker. Disease outcome of methylation groups was compared during follow-up. MI of all biomarkers was successfully obtained in 237 tumors of which 79 (33%) were classified as meth-N, 86 (36%) as meth-L, and 72 (30%) as meth-H. Meth-H status was a risk factor for distant recurrence (DR) (log-rank P = 0.007) and shorter disease-free survival (DFS) (log-rank P = 0.039). Methylation classification had strongest prognostic value for patients with ER-negative tumors. In multivariate analysis (n = 222), ER-negative meth-H patients had a 4.1-fold increased risk of DR (95% CI 1.80–9.59; meth-N HR 1.0, P = 0.001), a 4.2-fold increased risk of overall recurrence (OR) (95% CI 1.88–9.47; meth-N HR 1.0, P = 0.001), and a 3.1-fold shorter DFS (95% CI 1.57–5.98; meth-N HR 1.0, P = 0.003). Methylation classification of primary breast cancer is an independent prognostic factor for disease outcome in patients with ER-negative tumors. The study’s findings will have to be confirmed in an independent dataset.



Similar content being viewed by others
References
Brenton JD, Carey LA, Ahmed AA, Caldas C (2005) Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 23(29):7350–7360
Polyak K (2007) Breast cancer: origins and evolution. J Clin Invest 117(11):3155–3163
Sorlie T (2004) Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer 40(18):2667–2675
Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM (2006) The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics 7:96
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lonning PE, Borresen-Dale AL, Brown PO, Botstein D (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA (2005) Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet 365(9460):671–679
Mulero-Navarro S, Esteller M (2008) Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol 68(1):1–11
Kristensen LS, Nielsen HM, Hansen LL (2009) Epigenetics and cancer treatment. Eur J Pharmacol 625(1–3):131–142
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358(11):1148–1159
Lo PK, Sukumar S (2008) Epigenomics and breast cancer. Pharmacogenomics 9(12):1879–1902
de Maat MF, Umetani N, Sunami E, Turner RR, Hoon DS (2007) Assessment of methylation events during colorectal tumor progression by absolute quantitative analysis of methylated alleles. Mol Cancer Res 5(5):461–471
Feng W, Shen L, Wen S, Rosen DG, Jelinek J, Hu X, Huan S, Huang M, Liu J, Sahin AA, Hunt KK, Bast RC Jr, Shen Y, Issa JP, Yu Y (2007) Correlation between CpG methylation profiles and hormone receptor status in breast cancers. Breast Cancer Res 9(4):R57
Pasquali L, Bedeir A, Ringquist S, Styche A, Bhargava R, Trucco G (2007) Quantification of CpG island methylation in progressive breast lesions from normal to invasive carcinoma. Cancer Lett 257(1):136–144
Van der Auwera I, Bovie C, Svensson C, Trinh XB, Limame R, van Dam P, van Laere SJ, van Marck EA, Dirix LY, Vermeulen PB (2010) Quantitative methylation profiling in tumor and matched morphologically normal tissues from breast cancer patients. BMC Cancer 10:97
Fackler MJ, McVeigh M, Evron E, Garrett E, Mehrotra J, Polyak K, Sukumar S, Argani P (2003) DNA methylation of RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and invasive lobular breast carcinoma. Int J Cancer 107(6):970–975
Fackler MJ, McVeigh M, Mehrotra J, Blum MA, Lange J, Lapides A, Garrett E, Argani P, Sukumar S (2004) Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res 64(13):4442–4452
Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B (2005) Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin Cancer Res 11(6):2156–2162
de Maat MF, Narita N, Benard A, Yoshimura T, Kuo C, Tollenaar RA, de Miranda NF, Turner RR, van de Velde CJ, Morreau H, Hoon DS (2010) Development of sporadic microsatellite instability in colorectal tumors involves hypermethylation at methylated-in-tumor loci in adenoma. Am J Pathol 177(5):2347–2356
de Maat MF, van de Velde CJ, van der Werff MP, Putter H, Umetani N, Klein-Kranenbarg EM, Turner RR, van Krieken JH, Bilchik A, Tollenaar RA, Hoon DS (2008) Quantitative analysis of methylation of genomic loci in early-stage rectal cancer predicts distant recurrence. J Clin Oncol 26(14):2327–2335
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 96(15):8681–8686
Tanemura A, Terando AM, Sim MS, van Hoesel AQ, de Maat MF, Morton DL, Hoon DS (2009) CpG island methylator phenotype predicts progression of malignant melanoma. Clin Cancer Res 15(5):1801–1807
Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, Park SK, Noh DY, Yoo KY, Kang D (2010) Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet 19(21):4273–4277
Sunami E, Shinozaki M, Sim MS, Nguyen SL, Vu AT, Giuliano AE, Hoon DS (2008) Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res 10(3):R46
van Nes JG, de Kruijf EM, Faratian D, van de Velde CJ, Putter H, Falconer C, Smit VT, Kay C, van de Vijver MJ, Kuppen PJ, Bartlett JM (2011) COX2 expression in prognosis and in prediction to endocrine therapy in early breast cancer patients. Breast Cancer Res Treat 125(3):671–685
Hayes DF, Ethier S, Lippman ME (2006) New guidelines for reporting of tumor marker studies in breast cancer research and treatment: REMARK. Breast Cancer Res Treat 100(2):237–238
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 23(36):9067–9072
Umetani N, de Maat MF, Mori T, Takeuchi H, Hoon DS (2005) Synthesis of universal unmethylated control DNA by nested whole genome amplification with phi29 DNA polymerase. Biochem Biophys Res Commun 329(1):219–223
Putter H, Fiocco M, Geskus RB (2007) Tutorial in biostatistics: competing risks and multi-state models. Stat Med 26(11):2389–2430
Chang J, Hilsenbeck S (2010) Prognostic and predictive markers. In: Harris J, Lippman M, Morrow M, Osborne C (eds) Diseases of the breast, 4th edn. Lippincott Williams & Wilkins, Philadelphia, PA, pp 443–457
Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC Jr (2007) American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol 25(33):5287–5312
van Hoesel AQ, van de Velde CJ, Giuliano AE, Hoon DS (2008) Assessment of MINT 17 methylation in primary breast cancer and normal breast epithelia: a preliminary study. J Clin Oncol: Abstract 11113
Dunnwald LK, Rossing MA, Li CI (2007) Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast Cancer Res 9(1):R6
Castellano I, Allia E, Accortanzo V, Vandone AM, Chiusa L, Arisio R, Durando A, Donadio M, Bussolati G, Coates AS, Viale G, Sapino A (2010) Androgen receptor expression is a significant prognostic factor in estrogen receptor positive breast cancers. Breast Cancer Res Treat 124(3):607–617
Albain KS, Barlow WE, Shak S, Hortobagyi GN, Livingston RB, Yeh IT, Ravdin P, Bugarini R, Baehner FL, Davidson NE, Sledge GW, Winer EP, Hudis C, Ingle JN, Perez EA, Pritchard KI, Shepherd L, Gralow JR, Yoshizawa C, Allred DC, Osborne CK, Hayes DF (2010) Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol 11(1):55–65
Loi S, Haibe-Kains B, Desmedt C, Wirapati P, Lallemand F, Tutt AM, Gillet C, Ellis P, Ryder K, Reid JF, Daidone MG, Pierotti MA, Berns EM, Jansen MP, Foekens JA, Delorenzi M, Bontempi G, Piccart MJ, Sotiriou C (2008) Predicting prognosis using molecular profiling in estrogen receptor-positive breast cancer treated with tamoxifen. BMC Genomics 9:239
Mamounas EP, Tang G, Fisher B, Paik S, Shak S, Costantino JP, Watson D, Geyer CE Jr, Wickerham DL, Wolmark N (2010) Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol 28(10):1677–1683
Yau C, Benz CC (2008) Genes responsive to both oxidant stress and loss of estrogen receptor function identify a poor prognosis group of estrogen receptor positive primary breast cancers. Breast Cancer Res 10(4):R61
Albergaria A, Paredes J, Sousa B, Milanezi F, Carneiro V, Bastos J, Costa S, Vieira D, Lopes N, Lam EW, Lunet N, Schmitt F (2009) Expression of FOXA1 and GATA-3 in breast cancer: the prognostic significance in hormone receptor-negative tumours. Breast Cancer Res 11(3):R40
Ray PS, Wang J, Qu Y, Sim MS, Shamonki J, Bagaria SP, Ye X, Liu B, Elashoff D, Hoon DS, Walter MA, Martens JW, Richardson AL, Giuliano AE, Cui X (2010) FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res 70(10):3870–3876
Speers C, Tsimelzon A, Sexton K, Herrick AM, Gutierrez C, Culhane A, Quackenbush J, Hilsenbeck S, Chang J, Brown P (2009) Identification of novel kinase targets for the treatment of estrogen receptor-negative breast cancer. Clin Cancer Res 15(20):6327–6340
Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C (2007) An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol 8(8):R157
Berry DA, Cirrincione C, Henderson IC, Citron ML, Budman DR, Goldstein LJ, Martino S, Perez EA, Muss HB, Norton L, Hudis C, Winer EP (2006) Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer. JAMA 295(14):1658–1667
Colleoni M, Cole BF, Viale G, Regan MM, Price KN, Maiorano E, Mastropasqua MG, Crivellari D, Gelber RD, Goldhirsch A, Coates AS, Gusterson BA (2010) Classical cyclophosphamide, methotrexate, and fluorouracil chemotherapy is more effective in triple-negative, node-negative breast cancer: results from two randomized trials of adjuvant chemoendocrine therapy for node-negative breast cancer. J Clin Oncol 28(18):2966–2973
Ruike Y, Imanaka Y, Sato F, Shimizu K, Tsujimoto G (2010) Genome-wide analysis of aberrant methylation in human breast cancer cells using methyl-DNA immunoprecipitation combined with high-throughput sequencing. BMC Genomics 11:137
Bloushtain-Qimron N, Yao J, Snyder EL, Shipitsin M, Campbell LL, Mani SA, Hu M, Chen H, Ustyansky V, Antosiewicz JE, Argani P, Halushka MK, Thomson JA, Pharoah P, Porgador A, Sukumar S, Parsons R, Richardson AL, Stampfer MR, Gelman RS, Nikolskaya T, Nikolsky Y, Polyak K (2008) Cell type-specific DNA methylation patterns in the human breast. Proc Natl Acad Sci USA 105(37):14076–14081
Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, Eppenberger-Castori S, Vuaroqueaux V, Lerebours F, Welzel K, Maier S, Plum A, Niemann S, Foekens JA, Lesche R, Martens JW (2009) DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res 15(1):315–323
Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, Spyratos F, Thomssen C, Mueller V, Nahrig J, Schittulli F, Kates R, Lesche R, Schwope I, Kluth A, Marx A, Martens JW, Foekens JA, Schmitt M, Harbeck N (2007) DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients–Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer 43(11):1679–1686
Acknowledgments
This study was funded in part by Susan G. Komen Breast Cancer Foundation (Dave S.B. Hoon and Armando E. Giuliano), the Associates for Breast and Prostate Cancer Studies (Dave S.B. Hoon and Armando E. Giuliano), the Leslie and Susan Gonda (Goldschmied) Foundation (Dave S.B. Hoon), Avon Breast Foundation (Dave S.B. Hoon and Armando E. Giuliano), and Ruth and Martin H. Weil Fund (Dave S.B. Hoon).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
van Hoesel, A.Q., van de Velde, C.J.H., Kuppen, P.J.K. et al. Primary tumor classification according to methylation pattern is prognostic in patients with early stage ER-negative breast cancer. Breast Cancer Res Treat 131, 859–869 (2012). https://doi.org/10.1007/s10549-011-1485-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1485-3