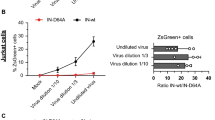
We studied the effect of KDM5 family demethylase inhibitors (JIB-04, PBIT, and KDOAM-25) on the penetration of SARS-CoV-2 pseudotyped viruses into differentiated Caco-2 cells and HEK293T cells with ACE2 hyperexpression. The above drugs were not cytotoxic. Only KDOAM-25 significantly reduced virus entry into the cells. The expression of ACE2 mRNA in Caco-2 significantly increased, while TMPRSS2 expression did not significantly change under these conditions. In differentiated Caco-2 cells, KDOAM-25 did not affect the expression of BRCA1, CDH1, TP53, SNAI1, VIM, and UGCG genes, for which an association with knockdown or overexpression of KDM5 demethylases or with the action of demethylase inhibitors had previously been shown. In undifferentiated Caco-2 cells, the expression of BRCA1, SNAI1, VIM, and CDH1 was significantly increased under the action of KDOAM-25.
Similar content being viewed by others
References
Laurie MT, Liu J, Sunshine S, Peng J, Black D, Mitchell AM, Mann SA, Pilarowski G, Zorn KC, Rubio L, Bravo S, Marquez C, Sabatino JJ, Mittl K, Petersen M, Havlir D, DeRisi J. SARS-CoV-2 variant exposures elicit antibody responses with differential cross-neutralization of established and emerging strains including Delta and Omicron. J. Infect. Dis. 2022;225(11):1909-1914. https://doi.org/10.1093/infdis/jiab635
Ashour NA, Abo Elmaaty A, Sarhan AA, Elkaeed EB, Moussa AM, Erfan IA, Al-Karmalawy AA. A systematic review of the global intervention for SARS-CoV-2 combating: from drugs repurposing to Molnupiravir approval. Drug Des. Devel. Ther. 2022;16:685-715. https://doi.org/10.2147/DDDT.S354841
Huang S, Fishell G. In SARS-CoV-2, astrocytes are in it for the long haul. Proc. Natl Acad. Sci. USA. 2022;119(30):e2209130119. https://doi.org/10.1073/pnas.2209130119
Knyazev E, Nersisyan S, Tonevitsky A. Endocytosis and transcytosis of SARS-CoV-2 across the intestinal epithelium and other tissue barriers. Front. Immunol. 2021;12:636966. https://doi.org/10.3389/fimmu.2021.636966
Nersisyan S, Shkurnikov M, Turchinovich A, Knyazev E, Tonevitsky A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PLoS One. 2020;15(7):e0235987. https://doi.org/10.1371/journal.pone.0235987
Tumber A, Nuzzi A, Hookway ES, Hatch SB, Velupillai S, Johansson C, Kawamura A, Savitsky P, Yapp C, Szykowska A, Wu N, Bountra C, Strain-Damerell C, Burgess-Brown NA, Ruda GF, Fedorov O, Munro S, England KS, Nowak RP, Schofield CJ, La Thangue NB, Pawlyn C, Davies F, Morgan G, Athanasou N, Müller S, Oppermann U, Brennan PE. Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem. Biol. 2017;24(3):371-380. https://doi.org/10.1016/j.chembiol.2017.02.006
Zheng YC, Chang J, Wang LC, Ren HM, Pang JR, Liu HM. Lysine demethylase 5B (KDM5B): A potential anti-cancer drug target. Eur. J. Med. Chem. 2019;161:131-140. https://doi.org/10.1016/j.ejmech.2018.10.040
Brewitz L, Tumber A, Pfeffer I, McDonough M.A, Schofield CJ. Aspartate/asparagine-β-hydroxylase: a high-throughput mass spectrometric assay for discovery of small molecule inhibitors. Sci. Rep. 2020;10(1):8650. https://doi.org/10.1038/s41598-020-65123-9
Betari N, Sahlholm K, Ishizuka Y, Teigen K, Haavik J. Discovery and biological characterization of a novel scaffold for potent inhibitors of peripheral serotonin synthesis. Future Med. Chem. 2020;12(16):1461-1474. https://doi.org/10.4155/fmc-2020-0127
Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, Babinski DJ, Frantz DE, Brekken RA, Quinn AM, Simeonov A, Easmon J, Martinez ED. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun. 2013;4:2035. https://doi.org/10.1038/ncomms3035
Chen M, Zhang XE. Construction and applications of SARS-CoV-2 pseudoviruses: a mini review. Int. J. Biol. Sci. 2021;17(6):1574-1580. https://doi.org/10.7150/ijbs.59184
Rosa RB, Dantas WM, do Nascimento JCF, da Silva MV, de Oliveira RN, Pena LJ. In vitro and in vivo models for studying SARS-CoV-2, the etiological agent responsible for COVID-19 pandemic. Viruses. 2021;13(3):379. https://doi.org/10.3390/v13030379.
Nikulin SV, Knyazev EN, Gerasimenko TN, Shilin SA, Gazizov IN, Zakharova GS, Poloznikov AA, Sakharov DA. Impedance spectroscopy and transcriptome analysis of choriocarcinoma BeWo b30 as a model of human placenta. Mol. Biol. (Mosk). 2019;53(3):467-475. https://doi.org/10.1134/S0026898419030133
Knyazev EN, Nyushko KM, Alekseev BY, Samatov TR, Shkurnikov MY. Suppression of ITGB4 gene expression in PC-3 cells with short interfering RNA induces changes in the expression of β-integrins associated with RGD-receptors. Bull. Exp. Biol. Med. 2015;159(4):541-545. https://doi.org/10.1007/s10517-015-3011-9
Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen. J. Biol. Chem. 2013;288(13):9408-9417. https://doi.org/10.1074/jbc.M112.419861
Kim MS, Cho HI, Yoon HJ, Ahn YH, Park EJ, ** YH, Jang YK. JIB-04, a small molecule histone demethylase inhibitor, selectively targets colorectal cancer stem cells by inhibiting the Wnt/β-catenin signaling pathway. Sci. Rep. 2018;8(1):6611. https://doi.org/10.1038/s41598-018-24903-0
Müller MR, Burmeister A, Skowron MA, Stephan A, Bremmer F, Wakileh GA, Petzsch P, Köhrer K, Albers P, Nettersheim D. Therapeutical interference with the epigenetic landscape of germ cell tumors: a comparative drug study and new mechanistical insights. Clin. Epigenetics. 2022;14(1):5. https://doi.org/10.1186/s13148-021-01223-1
Nersisyan SA, Shkurnikov MY, Osipyants AI, Vechorko VI. Role of ACE2/TMPRSS2 genes regulation by intestinal microRNA isoforms in the COVID-19 pathogenesis. Bulletin of RSMU. 2020;(2):16-18. https://doi.org/10.24075/brsmu.2020.024
Baddock HT, Brolih S, Yosaatmadja Y, Ratnaweera M, Bielinski M, Swift LP, Cruz-Migoni A, Fan H, Keown JR, Walker AP, Morris GM, Grimes JM, Fodor E, Schofield CJ, Gileadi O, McHugh PJ. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022;50(3):1484-1500. https://doi.org/10.1093/nar/gkab1303
Son J, Huang S, Zeng Q, Bricker TL, Case JB, Zhou J, Zang R, Liu Z, Chang X, Darling TL, Xu J, Harastani HH, Chen L, Gomez Castro MF, Zhao Y, Kohio HP, Hou G, Fan B, Niu B, Guo R, Rothlauf PW, Bailey AL, Wang X, Shi PY, Martinez ED, Brody SL, Whelan SPJ, Diamond MS, Boon ACM, Li B, Ding S. JIB-04 has broad-spectrum antiviral activity and inhibits SARS-CoV-2 replication and coronavirus pathogenesis. mBio. 2022;13(1):e0337721. https://doi.org/10.1128/mbio.03377-21
Beacon TH, Delcuve GP, Davie JR. Epigenetic regulation of ACE2, the receptor of the SARS-CoV-2 virus1. Genome. 2021;64(4):386-399. https://doi.org/10.1139/gen-2020-0124
Knyazev E, Maltseva D, Raygorodskaya M, Shkurnikov M. HIF-dependent NFATC1 activation upregulates ITGA5 and PLAUR in intestinal epithelium in inflammatory bowel disease. Front. Genet. 2021;12:791640. https://doi.org/10.3389/fgene.2021.791640
Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25(6):801-812. https://doi.org/10.1016/j.molcel.2007.03.001
Rajput K, Ansari MN, Jha SK, Medatwal N, Sharma P, Datta S, Kar A, Pani T, Cholke K, Rana K, Khan A, Mukherjee G, Deo SVS, Prabhu JS, Mukhopadhyay A, Bajaj A, Dasgupta U. RICTOR drives ZFX-mediated ganglioside biosynthesis to promote breast cancer progression. bioRxiv. 2022.01.10.475595. https://doi.org/10.1101/2022.01.10.475595
Bamodu OA, Huang WC, Lee WH, Wu A, Wang LS, Hsiao M, Yeh CT, Chao TY. Aberrant KDM5B expression promotes aggressive breast cancer through MALAT1 overexpression and downregulation of hsa-miR-448. BMC Cancer. 2016;16:160. https://doi.org/10.1186/s12885-016-2108-5
Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol. Cell. Biol. 2008;28(17):5312-5327. https://doi.org/10.1128/MCB.00128-08
Dabiri Y, Gama-Brambila RA, Taškova K, Herold K, Reuter S, Adjaye J, Utikal J, Mrowka R, Wang J, Andrade-Navarro MA, Cheng X. Imidazopyridines as potent KDM5 demethylase inhibitors promoting reprogramming efficiency of human iPSCs. iScience. 2019;12:168-181. https://doi.org/10.1016/j.isci.2019.01.012
Kuo KT, Huang WC, Bamodu OA, Lee WH, Wang CH, Hsiao M, Wang LS, Yeh CT. Histone demethylase JARID1B/KDM5B promotes aggressiveness of non-small cell lung cancer and serves as a good prognostic predictor. Clin. Epigenetics. 2018;10(1):107. https://doi.org/10.1186/s13148-018-0533-9
Schonfeld M, Averilla J, Gunewardena S, Weinman SA, Tikhanovich I. Male-specific activation of lysine demethylases 5B and 5C mediates alcohol-induced liver injury and hepatocyte dedifferentiation. Hepatol. Commun. 2022;6(6):1373-1391. https://doi.org/10.1002/hep4.1895
Nikulin SV, Mnafki (Krainova) NA, Shilin SA, Gazizov IN, Maltseva DV. Ribosome inactivation and the integrity of the intestinal epithelial barrier. Mol. Biol. (Mosk). 2018;52(4):675-682. https://doi.org/10.1134/S0026898418040146
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Kletochnye Tekhnologii v Biologii i Meditsine, No. 1, pp. 40-46, March, 2023
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Knyazev, E.N., Kalinin, R.S., Abrikosova, V.A. et al. KDM5 Family Demethylase Inhibitor KDOAM-25 Reduces Entry of SARS-CoV-2 Pseudotyped Viral Particles into Cells. Bull Exp Biol Med 175, 150–156 (2023). https://doi.org/10.1007/s10517-023-05827-w
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10517-023-05827-w