Abstract
Carbapenem-resistant Klebsiella pneumoniae (CRKP), as one of the most common drug-resistant bacteria threatening human health, is hyper-resistant to multiple antimicrobial drugs and carbapenems, which can be dealt with only limited clinical treatment options. This study described the epidemiological characteristics of CRKP in this tertiary care hospital from 2016 to 2020. Specimen sources included blood, sputum, alveolar lavage fluid, puncture fluid, secretions from a burn wound, and urine. Among the 87 carbapenem-resistant strains, ST11 was the predominant isolate, followed by ST15, ST273, ST340, and ST626. These STs were in broad agreement with the STs defined by pulsed-field gel electrophoresis clustering analysis in discriminating clusters of related strains. Most CRKP isolates contained the blaKPC-2 gene, some isolates carried the blaOXA-1, blaNDM-1, and blaNDM-5 genes, and the isolates carrying carbapenem resistance genes were more resistant to the antimicrobials of β-lactams, carbapenems, macrolides, and fluoroquinolone. The OmpK35 and OmpK37 genes were detected in all CRKP strains, and the Ompk36 gene was detected in some CRKP strains. All detected OmpK37 had 4 mutant sites, and OmpK36 had 11 mutant sites, while no mutant sites were found in OmpK35. More than half of the CRKP strains contained the OqxA and OqxB efflux pump genes. The virulence genes were most commonly combined with urea-wabG-fimH-entB-ybtS-uge-ycf. Only one CRKP isolate was detected with the K54 podoconjugate serotype. This study elucidated the clinical epidemiological features and molecular ty** of CRKP, and grasped the distribution of drug-resistant genotypes, podocyte serotypes, and virulence genes of CRKP, providing some guidance for the subsequent treatment of CRKP infection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Klebsiella pneumoniae is a gram-negative bacterium that is widely distributed in the environment such as surface water, sewage, soil, and plants, and it may also exist in the intestinal tract and nasopharynx of humans (Podschun and Ullmann 1998). CRKP is an increasingly important pathogen of hospital-acquired infections that may cause a wide range of infections, including urinary tract infections, pneumonia, bacteremia, liver abscesses, and brain abscesses, and may be life-threatening in severe infections (Paczosa and Mecsas 2016). In clinical practice, great success has been achieved in the treatment of pathogenic infections with antimicrobials, but in recent years, K. pneumoniae has rapidly developed resistance to all known antimicrobials, including carbapenems, and this phenomenon can be seen all over the world. Due to its highly transmissible nature, the emergence of carbapenem-resistant K. pneumoniae in endemic areas of Klebsiella pneumoniae may lead to a dangerous local increase in the number of carbapenem-resistant K. pneumoniae, and the emergence of carbapenem-resistant K. pneumoniae has clearly become a world public health problem that may be faced by all the human beings (Holden et al. 2018; Doi 2019; Karampatakis et al. 2016). The main resistance mechanisms of carbapenem-resistant K. pneumoniae are currently shown as follows: (1) K. pneumoniae can be prevented by encoding genes (e.g., New Delhi metallo-β-lactamase [NDM], K. pneumoniae carbapenemase [KPC], Verona integron-encoded metallo-β-lactamase [VIM], oxacillin-hydrolyzing β-lactamase [OXA], and imipenem-hydrolyzing β-lactamase [IMP]) to hydrolyze carbapenemases produced by the carbapenemase, which is an enzyme with a positive effect on the hydrolysis of carbapenems. Carbapenemases are catalytically efficient endocannabinoid enzymes that may result in elevated minimum inhibitory concentrations of carbapenem and include classes A, B, and D. The three major families of class-A serine carbapenemases include NMC/IMI, SME, and KPC enzymes; the most common families of class-B metallo-lactamases include blaNDM, blaVIM, blaIMP, blaGIM, and blaSIM enzymes; class-D serine carbapenemase includes blaOXA β-lactamase. (2) The expression of efflux pump gene allows bacterial cells to actively and rapidly squeeze out carbapenems, avoiding the bacterial cell wall from being damaged by carbapenems. (3) The permeability of the outer membrane is reduced through the production of β-lactamases (AmpC) and the alterations in the bacterial cell membrane (pore protein mutations in OmpK35 and OmpK36), thus blocking carbapenem drugs from entering into the bacteria (Alizadeh et al. 2018; Durante-Mangoni et al. 2019; Girlich et al. 2019; Queenan and Bush 2007). The synthesis of carbapenemase-encoding genes has caused numerous difficulties for clinicians, and the drugs do not work as well as they should, making it difficult to develop treatment plans for patients. The common carbapenemase-encoding genes in China are mainly KPC, NDM, and IMP n (Zhang et al. 2020a). A weak correlation exists between the expression of carbapenem antimicrobial resistance in CRKP and the virulence genes they possess, and these strains may carry multiple virulence genes that may have the ability to encode capsules (magA, wcaG), express high viscosity, mucus phenotype regulators (magA, rpmA), produce mycobacterial hair adhesion elements (fimH), synthesize lipopolysaccharides (wabG, uge, ycf), and construct iron acquisition systems (iutA, iroN, entB, KfuB), iron carriers (aerobactin), associated with allantoin metabolism (allS), yersin bacteriocin (ybtS), and other virulence factors (vatD); the functions possessed by these virulence genes allow them to overcome host defenses (Hartman et al. 2009; Yu et al. 2008, 2006, 2007; Remya et al. 2019; Candan and Aksöz, 2015). Almost all CRKPs prevalent worldwide are multi-locus sequence ty** (ST) 258 and its related variants; for example, ST11, which is widely prevalent in China, differs from ST258 in only one gene locus, but it is not yet clear why ST258 and its related variants spread so widely and successfully (Qi et al. 2011; Chen et al. 2014).
In this study, the antimicrobial susceptibility test was performed on 87 carbapenem-resistant K. pneumoniae strains. Multi-locus sequence ty**, drug resistance genes, virulence genes, and capsular serotypes were detected using PCR amplification; clonal relationship detection was performed using pulsed-field gel electrophoresis (PFGE) technique to clarify the molecular epidemiological characteristics of CRKP in this hospital.
Materials and methods
Sample collection and bacterial isolates
A total of 87 non-replicated carbapenem-insensitive K. pneumoniae strains were collected from January 2016 to December 2020 in a tertiary care hospital in Shijiazhuang, Hebei Province, China. All isolates of specimens (sputum, secretions, urine, alveolar lavage, puncture fluid, and blood) were obtained from the clinical departments of this tertiary hospital, and there was no restriction on age, sex, disease type, and clinical department of the patients who provided the specimens. Eighty-seven isolates were identified using a MALDI-TOF mass spectrometer (AUTOFLEXSpeed, Germany). The MIC values of imipenem (MIC ≥ 4 μg/mL), meropenem (MIC ≥ 4 μg/mL), and ertapenem (MIC ≥ 2 μg/mL) were determined using the microbroth dilution method with a fold reference (CLSI M100, 2021). According to the guidelines in the Clinical and Laboratory Standards Institute (CLSI) (2019, 29), the inclusion criteria for the guidelines were reduced susceptibility to at least one carbapenem (imipenem, meropenem, and ertapenem) as carbapenem resistance. All 87 isolates were stored in a − 80℃ refrigerator.
Antimicrobial sensitivity experiments
Antimicrobial susceptibility testing is conducted using the microdilution method (MIC method) according to the manufacturer ‘s instructions. To put it simply, a fresh bacterial strain is prepared into a 0.5 mcf suspension, added to a 96-well plate following the instructions, and incubated in a constant temperature incubator for 24 h. The number of negative wells for each antimicrobial is then recorded and entered into the result analysis system provided by the manufacturer to obtain the corresponding antimicrobial ‘s minimum inhibitory concentration. Finally, these figures are entered into the manufacturer ‘s result analysis system to obtain the minimum inhibitory concentration. Thirty-one antimicrobials used in antimicrobial trials included ampicillin, ampicillin-sulbactam, tetracycline, chloramphenicol, trimethoprim-sulfamethoxazole, cefazolin, cefotaxime, ceftazidime, cefoxitin, gentamicin, imipenem, Nalidixan, azithromycin, sulfisoxazole, ciprofloxacin, amoxicillin-clavulanate, cefotaxime-clavulanate, ceftazidime-clavulanate, colistin, polymyxin B, minocycline, amikacin, aztreonam, cefepime, meropenem, ertapenem levofloxacin, doxycycline, kanamycin, streptomycin, and gemifloxacin. The breakpoint of measured antimicrobial MIC values was obtained according to the corresponding standards of the American Committee for Clinical Laboratory Standardization (CLSI M100, 2021) for the corresponding sensitive (S), moderately sensitive (I), and resistant (R) results. The antimicrobial sensitivity test was performed using E.coli ATCC25922 and K. pneumoniae ATCC700603 as the reference standard.
Drug resistance gene, capsular serotype, and virulence gene detection
The polymerase chain reaction was performed using specific primer sequences with TaKaRa ‘s Premix Taq configuration system using PTC-200 and Veriti. Twelve resistance genes (blaKPC, blaNDM, blaVIM, blaOXA-48, blaOXA-1-like, blaIMP, blaGES, blaSHV, blaCTX-M-65, ompK35, ompK36, ompK37), 16 common virulence genes (WcaG, ureA, wabG, fimH, entB, iutA, ybtS, rmpA, IroN, KfuB, aerobactin, alls, uge, vatD, magA, and ycf), and seven capsular serotype genes (K1, K2, K3, K5, K20, K54, and K57) were selected (Kaczmarek et al. 2006; Dallenne et al. 2010; Zhan et al. 2017; Zhang et al. 2018). The reaction program for the total reaction system of 25 μL was set as follows: pre-denaturation temperature 94 °C for 2 min, denaturation temperature 94 °C for 20 s, annealing temperature depending on different primers (blaKPC, blaVIM, blaIMP, ompK35, ompK36, and ompK37 55 °C; blaGES and blaOXA-48 57 °C; blaOXA-1-like and uge 60 °C; blaSHV, K20, entB, and ybtS 56 °C; blaCTX-M-65, blaNDM, K1, and WcaG 54 °C; ureA, wabG, ycf, rmpA, aerobactin, IroN, KfuB, alls, K3, K5, K54, and K57 58 °C; K2 64 °C; fimH 62 °C; iutA 66 °C; vatD and magA 48 °C) (Kaczmarek et al. 2006; Dallenne et al. 2010; Zhan et al. 2017; Zhang et al. 2018), time 30 s, extension temperature 72℃, number of amplification cycles 35, final extension temperature 72℃, time 5 min. PCR products were analyzed using a fully automated electrophoresis analyzer (QIAXEL) and Sanger sequencing was performed (sent to Bei**g DynaBio), and the sequencing results were analyzed using NCBI BLAST (https://blast.ncbi.nlm.nih.gov/blast.cgi) for comparative analysis.
Detection of efflux pump gene
7500 Real-Time PCR System (Applied Biosystems) was used to detect OqxA and OqxB efflux pump genes, primers were synthesized by Bei**g DynaBio, the primer system used was TaKaRa ‘s Premix Ex TaqTM (Probe qPCR), and the amplification procedure was as follows: initial incubation at 95 °C for 2 min, followed by 40 cycles of 10 s at 95 °C, 30 s at 60 °C, and 10 s at 72 °C (Zhong et al. 2014). The strains that tested positive for DNA were extracted and reverse-transcribed into cDNA using TaKaRa ‘s PrimeScript™ II 1st Strand cDNA Synthesis Kit s. The strains were then configured using the Premix Ex Taq™ system and subjected to real-time fluorescent quantitative PCR. Amplification conditions are as described above. The 2^-ΔΔCT of each strain was calculated as the relative expression of the efflux pump gene for each strain, using the rpoB gene as the reference gene and strain number 20021762; the CRKP strain as the reference. Each sample was processed in triplicate.
Multi-locus sequence ty** (MLST)
PCR amplification of seven housekee** genes of K. pneumoniae (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) was also performed using TaKaRa ‘s Premix Taq configuration system, polymerase chain reaction using PTC-200 and Veriti, and total reaction system 50 μL. The reaction program was set as follows: pre-denaturation at 94 °C for 2 min, denaturation at 94 °C for 20 s, annealing temperature depending on different primers (gapA 60 °C, TonB 45 °C, infB, mdh, pgi, phoE, and rpoB all 50 °C) for 30 s, extension at 72 °C for 30 s, number of amplification cycles 35, final extension at 72 °C for 5 min. The PCR products were analyzed using a fully automated electrophoresis analyzer (QIAXEL), and Sanger sequencing was performed (sent to Bei**g DynaScience Biotech). The sequencing results were typed for STs using the BIGSdb-Pasteur database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
Pulsed-field gel electrophoresis (PFGE)
PFGE assays were performed on all collected carbapenem-resistant K. pneumoniae strains. Briefly, the genomic DNA of carbapenem-resistant K. pneumoniae was prepared by embedding K. pneumoniae cells in Seakem Gold Agarose and then digesting them using XbaI enzymes at 37 °C for 4 h. The genomic DNA of carbapenem-resistant K. pneumoniae was then stained using a Bio-Rad CHEF III system (120° angle, 6 V/cm, switching time 6 s, 36 s) at 14 °C for 18.5 h. After completion of electrophoresis, DNA bands were stained using GelRed and imaged with a gel imaging system (BIO-RAD Molecular Imager® Gel DocTM XR + with Image LabTM software). Salmonella serovar Brendaup strain H9812 was used as a molecular marker. DNA profiles were interpreted using the criteria of Tenover et al. (1995) (Zhan et al. 2017). PFGE patterns were analyzed using Bionumerics software (Applied Mathematics, Sint-Martens-Latem, Belgium) using dice similarity coefficients. The isolates with a similarity of more than 80% were defined as identical PFGE clusters.
Results
Sample types and sources of strains isolated from K. pneumonia
The number of CRKPs isolated per year from 2016 to 2020 in this tertiary care hospital was 14, 5, 6, 28, and 34 strains, respectively. The vast majority of the isolated CRKP specimens were originated from sputum 78.16% (68/87), followed by burn secretions 9.20% (8/87), urine 3.45% (3/87), alveolar lavage fluid 3.45% (3/87), puncture fluid 3.45% (3/87), and blood 2.30% (2/87). The detection rate of CRKP in the lower respiratory tract was high; CRKP was also widely distributed in clinical departments of hospitals, with the majority of carbapenem-resistant K. pneumoniae coming from the respiratory medicine department 21.84% (19/87), intensive care unit 21.84% (19/87), burns department 16.09% (14/87), rehabilitation department 13.79% (12/87), geriatrics department 9.20% (8/87), neurosurgery department 4.60% (4/87), thoracic surgery department 2.30% (2/87), nephrology department 2.30% (2/87), hematology department 2.30% (2/87), general surgery department 2.30% (2/87), neurology department 1.15% (1/87), gastroenterology department 1.15% (1/87), and endocrinology department 1.15% (1/87). Respiratory and Critical Care Medicine remain the main clinical departments prevalent with carbapenem-resistant K. pneumoniae. Although there are relatively fewer carbapenem-resistant K. pneumoniae in other departments, there is a small amount of CRKP epidemic transmission.
Antibacterial drugs, carbapenem-resistant genes, and efflux pump genes
Thirty-one antimicrobials commonly used in clinical practice were selected according to the microbroth dilution method (MIC method), which was developed according to the manufacturer ‘s method. Ampicillin, ampicillin-sulbactam, ceftazidime clavulanic acid, ceftazidime clavulanic acid, cefazolin, ceftazidime, cefoxitin, cefepime, amoxicillin-clavulanic acid, gemifioxacin, levofloxacin, ciprofloxacin, nalidixic acid, meropenem, imipenem, and azithromycin are antimicrobial drugs with a 100% resistance rate. The resistance rate is 98.85% for amineptide and ertapenem. The resistance rates for gentamicin, kanamycin, sulfisoxazole, trimethoprim, amikacin, chloramphenicol, tetracycline, doxycycline, streptomycin, and minocycline were 90.80%, 89.66%, 79.31%, 77.01%, 72.41%, 71.59%, 70.11%, 66.67%, 21.84%, and 10.31%. Colistin and polymyxin B’s resistance rate is 3.45%. The specific MIC values of the 31 antimicrobials are shown in the attached information. The resistance rate of CRKP to 20 antimicrobials in 31 antimicrobial sensitivity assays exceeded 90%, with an alarming 100% resistance rate of CRKP to 17 antimicrobials. All CRKPs were multi-drug-resistant (MDR), with one strain being insensitive to β-lactams, chloramphenicol, macrolides, fluoroquinolones, aminoglycosides, sulfonamides, tetracyclines, carbapenems, and glycopolypeptide antimicrobials. The distribution of resistant genotypes among 87 carbapenem-resistant K. pneumoniae strains was blaKPC-2 type 96.55% (84/87), blaOXA-1 family group 14.94% (13/87), blaNDM-1 type 2.30% (2/87), and blaNDM-5 type 1.15% (1/87), while blaVIM, blaIMP, and blaOXA-48 resistance genes were not detected; the extended-spectrum-beta-lactamase genes blaSHV 95.40% (83/87), blaCTX-M-65 75.86% (66/87); the outer membrane genes OmpK35 100% (87/87), OmpK36 22.99% (20/87), and OmpK37 100% (87/87); and the efflux pump genes OqxA 63.22% (55/87) and OqxB 63.22% (55/87). The detailed distribution of drug resistance genes is shown in Table 1. No mutation sites were found in all detected CRKPs of OmpK35, while all detected OmpK37 genes had four mutation types of p.I70M, p.I128M, m233_None234insQ, and p.N230G. The GenBank ID of the pore protein sequence of the reference Klebsiella pneumoniae strain of OmpK37 was AJ011502. Eleven types of mutation were also detected in the OmpK36 gene, including p.F207W, p.D224E, p.L59V, p.L228V, p.N49S, p.Q227_None679del, p.L191S, p.A190_None568del, p.N218H, p. E232R, and p.A217S. The GenBank accession number of the pore protein sequence of the Klebsiella pneumoniae strain referenced by OmpK36 was Z33506.1. Detailed results of carbapenem resistance genes, outer membrane protein genes, efflux pump genes, extended-spectrum-beta-lactamase genes, and antimicrobial sensitivity test of each isolated strain are shown in Fig. 1. We selected 19 CRKP strains of KPC-2 type for mRNA expression assay of exocytosis pump gene. Expression of mRNA for the efflux pump gene was detected in all 19 CRKP strains. The relative expression of the efflux pump genes is detailed in Fig. 2.
Capsular serotypes and virulence genes
The results were verified by amplification of specific primers, the amplified products were verified by electrophoresis, and the products verified by electrophoresis were sent to the company for sequencing, and the sequencing results were verified by BLAST. Only K54 1.15% (1/87) of the 87 isolated carbapenem-resistant K. pneumoniae were detected in the capsular serotype, whereas K1, K2, K3, K5, K20, and K57 capsular serotypes were not detected. Virulence genes were detected in 100.00% (87/87) of WabG, 100.00% (87/87) of ycf, 100.00% (87/87) of entB, 94.25% (82/87) of urea, 91.95% (80/87) of ybtS, 88.51% (77/87) of uge, and 83.91% (73/87) of fimH. Detailed distribution of virulence genes is shown in Table 1. The most common combination of virulence genes in this study was urea-wabG-fimH-entB-ybtS-uge-ycf; the virulence gene combination in the CRKP strain with capsular serotype K54 was WcaG-urea-wabG-fimH-entB-KfuB-alls-uge-ycf. The detailed results are shown in Fig. 3.
Molecular characteristics of K. pneumoniae isolates
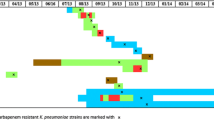
Among the 87 CRKP strains, five STs were identified, including ST11 93.10% (81/87), ST15 3.45% (3/87), ST273 1.15% (1/87), ST340 1.15% (1/87), and ST626 1.15% (1/87); the results of PFGE clustering analysis showed that 87 CRKP strains could be divided into five clusters: cluster A comprised of ST15 3.45% (3/87), cluster B comprised of ST11 10.34% (9/87), cluster C comprised of ST11 55.17% (48/87), cluster D comprised of ST11 4.60% (4/87), cluster E comprised of ST11 18.39% (16/87), and ST273 1.15% (1/87). The other six CRKP strains were not clustered with other strains, including ST11 4.60% (4/87), ST626 1.15% (1/87), and ST340 1.15% (1/87). The detailed results are shown in Fig. 4.
Pulse-field gel electrophoresis results of 87 strains of CRKP. Note: the red line is the dividing line with 80% similarity, the light green box represents cluster A; the sky blue box represents cluster B; the pink box represents cluster C; the light yellow box represents cluster D; the dark blue box represents the E cluster
Discussion
The globally first case of K. pneumoniae type blaKPC was discovered in the USA in 1996. Over time, carbapenem-resistant bacteria capable of producing blaKPC have spread to most areas in the USA, Israel, and southern European countries (especially Greece and Italy), as well as to the South American continent and China (Munoz-Price et al. 2013). The first blaKPC-2 strain of K. pneumoniae was discovered in Zhejiang Province of China in 2007, and since then blaKPC-2 pneumoniae has spread rapidly to other provinces and cities such as Jiangsu (Qi et al. 2011), Anhui (Qi et al. 2011), Fujian (Chen et al. 2005). While in the USA and European countries, K. pneumoniae ST258 contributes significantly to the spread of carbapenem resistance, in the Chinese region, ST11 dominates; ST11 is a single motif variant (TonB) of ST258 and they are closely related. It was reported that ST11 and ST258 belonged to the clonal complex of CC258, which included ST11, ST258, and five other STSs (ST270, ST340, ST379, ST407, and ST418) (Qi et al. 2011; Wang et al. 2018). The CRKP isolated from this tertiary care hospital had a predominant ST11 type, and the number of ST11 type of CRKP in this tertiary care hospital showed a significant annual increase over the 5-year period from 2016 to 2020. This is consistent with the results of other Chinese researchers (Zhao et al. 2021; Hu et al. 2020). All CRKPs could be clearly divided into 5 taxa in the PFGE clustering analysis, and taxon A was full of ST15 clonal CRKP but had a low number of 3 strains compared to the results of a study in a medical center in northeastern China (Chen et al. 2021). CRKP of taxa B, C, D, and E were all dominated by ST11, but there were still some differences even among the same ST11 clonotype, which was probably because multiple incompatibilities between plasmids could lead to different resistance genes carried on the plasmids of each isolate, thus resulting in individual differences (Kim et al. 2021).
It was found in the virulence gene test results that virulence genes were more abundant in some CRKPs compared with the results of other researchers, especially the detection rate of rmpA, magA, and iutA genes was significantly higher than that of isolates from a tertiary hospital in Chongqing area (Zeng et al. 2021), but the results were closer to those of a tertiary care hospital in the Zhejiang area (Zhan et al. 2017). The rmpA, magA, and iutA virulence genes tend to be common in HVKP, which may be a sign that the CRKP within this hospital is evolving toward high virulence. The virulence plasmids which were considered non-conjugated in most previous studies are present only in HVKP. Now there has been a worldwide spread of virulence plasmids from HVKP into CRKP, in which the plasmids are increasingly associated with the spread of virulence factors, leading to the spread of important pathogenic characteristics, with CRKP possessing highly virulent plasmids being considerably more pathogenic than before (Kopotsa et al. 2020; Russo et al. 2014; Zhang et al. 2020b). Interestingly, a blaNDM-1 carbapenemase-type CRKP strain had capsular serotype K54, which was the first time in the region that a CRKP isolate was found to have capsular serotype K54; however, according to related reports, K54 was one of the supervirulent capsular serotypes of K. pneumoniae (Turton et al. 2018). The virulence gene combination of this blaNDM-1 isolate was WcaG-urea-wabG-fimH-entB-KfuB-alls-uge-ycf with two additional virulence genes of KfuB-alls compared with the virulence gene combinations of other isolates; the results of the correlation study concluded that there was a strong correlation between kfuB, alls, and K1 capsular serotype isolates, and that all K1 strains were considered positive for kfuB and alls (Yu et al. 2008). However, the podosome serotype K54 of the CRKP strain isolated in this study possessed the combination of KfuB-alls, which is a very special finding. The virulence plasmid expressing K54 serotype might be accidentally obtained during the transmission of this isolate, which meant that this isolate had both high resistance of ST11 and high virulence of K54. Subsequent studies may be conducted to verify the plasmid horizontal transfer ability of this strain and to obtain the complete genetic information of its plasmid to determine the cause of this phenomenon (Yang et al. 2021).
The resistance genotypes of CRKP collected during our study were mainly the prevalent genotype of blaKPC-2, with blaNDM-1 and blaNDM-5 resistance genotypes accounting for very few proportions. The types of outer membrane proteins were dominated by OmpK35 and OmpK37. The vast majority of isolates contained both OqxA and OqxB efflux pump genes. ST types were mainly ST11, with ST15, ST273, ST626, and ST340 accounting for a few proportions. The strains with blaKPC and blaNDM resistant genotypes showed very strong resistance to common antimicrobials, and there were also individuals with abundant virulence genes in these CRKPs, which means that some CRKP strains may gradually become novel K. pneumoniae with mixed characteristics of super-resistance and high virulence, so it is a very necessary task to timely monitor, prevent, and control CRKP epidemic disease in hospitals.
Data availability
The original contributions presented in the study are included in the article/Supplementary Material; further inquiries can be directed to the corresponding authors.
References
Aanensen D, Spratt B (2005) The multilocus sequence ty** network: mlst.net. Nucleic Acids Res 33(Web Server issue):W728-733. https://doi.org/10.1093/nar/gki415
Alizadeh N, Rezaee M, Kafil H, Barhaghi M, Memar M, Milani M et al (2018) Detection of carbapenem-resistant Enterobacteriaceae by chromogenic screening media. J Microbiol Methods 153:40–44. https://doi.org/10.1016/j.mimet.2018.09.001
Candan ED, Aksöz N (2015) Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol 62(4):867–874. https://doi.org/10.18388/abp.2015_1148
Chen L, Mathema B, Chavda K, DeLeo F, Bonomo R, Kreiswirth B (2014) Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22(12):686–696. https://doi.org/10.1016/j.tim.2014.09.003
Chen D, Li H, Zhao Y, Qiu Y, **ao L, He H et al (2020) Characterization of carbapenem-resistant Klebsiella pneumoniae in a tertiary hospital in Fuzhou. China J Appl Microbiol 129(5):1220–1226. https://doi.org/10.1111/jam.14700
Chen J, Hu C, Wang R, Li F, Sun G, Yang M et al (2021) Shift in the dominant sequence type of carbapenem-resistant Klebsiella pneumoniae bloodstream infection from ST11 to ST15 at a medical center in Northeast China, 2015–2020. Infect Drug Resist 14:1855–1863. https://doi.org/10.2147/idr.S311968
Cuzon G, Naas T, Truong H, Villegas M, Wisell K, Carmeli Y et al (2010) Worldwide diversity of Klebsiella pneumoniae that produce beta-lactamase blaKPC-2 gene. Emerg Infect Dis 16(9):1349–1356. https://doi.org/10.3201/eid1609.091389
Dallenne C, Da Costa A, Decré D, Favier C, Arlet G (2010) Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65(3):490–495. https://doi.org/10.1093/jac/dkp498
Doi Y (2019) Treatment options for carbapenem-resistant gram-negative bacterial infections. Clin Infect Dis 69(Suppl 7):S565-s575. https://doi.org/10.1093/cid/ciz830
Doménech-Sánchez A, Hernández-Allés S, Martínez-Martínez L, Benedí V, Albertí S (1999) Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in beta-lactam antibiotic resistance. J Bacteriol 181(9):2726–2732. https://doi.org/10.1128/jb.181.9.2726-2732.1999
Doménech-Sánchez A, Pascual A, Suárez A, Alvarez D, Benedí V, Martínez-Martínez L (2000) Activity of nine antimicrobial agents against clinical isolates of Klebsiella pneumoniae producing extended-spectrum beta-lactamases and deficient or not in porins. J Antimicrob Chemother 46(5):858–859. https://doi.org/10.1093/jac/46.5.858
Durante-Mangoni E, Andini R, Zampino R (2019) Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect 25(8):943–950. https://doi.org/10.1016/j.cmi.2019.04.013
Girlich D, Grosperrin V, Naas T, Dortet L (2019) CHROMagar™ ESBL/mSuperCARBA bi-plate medium for detection of ESBL- and carbapenemase-producing Enterobacteriaceae from spiked stools. Diagn Microbiol Infect Dis 95(2):107–112. https://doi.org/10.1016/j.diagmicrobio.2019.05.002
Han R, Shi Q, Wu S, Yin D, Peng M, Dong D et al (2020) Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol 10:314. https://doi.org/10.3389/fcimb.2020.00314
Hartman L, Selby E, Whitehouse C, Coyne S, Jaissle J, Twenhafel N et al (2009) Rapid real-time PCR assays for detection of Klebsiella pneumoniae with the rmpA or magA genes associated with the hypermucoviscosity phenotype: screening of nonhuman primates. J Mol Diagn 11(5):464–471. https://doi.org/10.2353/jmoldx.2009.080136
Hernández-Allés S, Albertí S, Álvarez D, Doménech-Sánchez A, Martínez-Martínez L, Gil J, Benedí V (1999) Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology (reading) 145(Pt 3):673–679. https://doi.org/10.1099/13500872-145-3-673
Holden V, Wright M, Houle S, Collingwood A, Dozois C, Adams M, Bachman M (2018) Iron acquisition and siderophore release by carbapenem-resistant sequence type 258 Klebsiella pneumoniae. mSphere 3(2). https://doi.org/10.1128/mSphere.00125-18
Hu Y, Liu C, Shen Z, Zhou H, Cao J, Chen S et al (2020) Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect 9(1):1771–1779. https://doi.org/10.1080/22221751.2020.1799721
Huang W, Zhang J, Zeng L, Yang C, Yin L, Wang J et al (2021) Carbapenemase production and epidemiological characteristics of carbapenem-resistant Klebsiella pneumoniae in Western Chongqing, China. Front Cell Infect Microbiol 11:775740. https://doi.org/10.3389/fcimb.2021.775740
Huang Y, Rana A, Wenzler E, Ozer E, Krapp F, Bulitta J et al (2022) Aminoglycoside-resistance gene signatures are predictive of aminoglycoside MICs for carbapenem-resistant Klebsiella pneumoniae. J Antimicrob Chemother 77(2):356–363. https://doi.org/10.1093/jac/dkab381
** X, Chen Q, Shen F, Jiang Y, Wu X, Hua X et al (2021) Resistance evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae ST11 during treatment with tigecycline and polymyxin. Emerg Microbes Infect 10(1):1129–1136. https://doi.org/10.1080/22221751.2021.1937327
Kaczmarek F, Dib-Hajj F, Shang W, Gootz T (2006) High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of bla(ACT-1) beta-lactamase production, porin OmpK35/36 insertional inactivation, and down-regulation of the phosphate transport porin phoe. Antimicrob Agents Chemother 50(10):3396–3406. https://doi.org/10.1128/aac.00285-06
Karampatakis T, Antachopoulos C, Iosifidis E, Tsakris A, Roilides E (2016) Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Future Microbiol 11:809–823. https://doi.org/10.2217/fmb-2016-0042
Kim J, Cho Y, Choi J, Wi Y, Ko K (2021) Two distinct genotypes of KPC-2-producing Klebsiella pneumoniae isolates from South Korea. Antibiotics (Basel) 10(8). https://doi.org/10.3390/antibiotics10080911
Kopotsa K, Mbelle N, Osei Sekyere J (2020) Epigenomics, genomics, resistome, mobilome, virulome and evolutionary phylogenomics of carbapenem-resistant Klebsiella pneumoniae clinical strains. Microb Genom 6(12). https://doi.org/10.1099/mgen.0.000474
Li J, Zhang H, Ning J, Sajid A, Cheng G, Yuan Z, Hao H (2019) The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control 8:44. https://doi.org/10.1186/s13756-019-0489-3
Munoz-Price L, Poirel L, Bonomo R, Schwaber M, Daikos G, Cormican M et al (2013) Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis 13(9):785–796. https://doi.org/10.1016/s1473-3099(13)70190-7
Nakano R, Nakano A, Hikosaka K, Kawakami S, Matsunaga N, Asahara M et al (2014) First report of metallo-β-lactamase NDM-5-producing Escherichia coli in Japan. Antimicrob Agents Chemother 58(12):7611–7612. https://doi.org/10.1128/aac.04265-14
Paczosa M, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80(3):629–661. https://doi.org/10.1128/mmbr.00078-15
Podschun R, Ullmann U (1998) Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, ty** methods, and pathogenicity factors. Clin Microbiol Rev 11(4):589–603. https://doi.org/10.1128/cmr.11.4.589
Qamar M, Walsh T, Toleman M, Tyrrell J, Saleem S, Aboklaish A et al (2019) Dissemination of genetically diverse NDM-1, -5, -7 producing-gram-negative pathogens isolated from pediatric patients in Pakistan. Future Microbiol 14:691–704. https://doi.org/10.2217/fmb-2019-0012
Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y (2011) ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66(2):307–312. https://doi.org/10.1093/jac/dkq431
Queenan A, Bush K (2007) Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20(3):440–458. https://doi.org/10.1128/cmr.00001-07. (table of contents)
Remya P, Shanthi M, Sekar U (2019) Characterisation of virulence genes associated with pathogenicity in Klebsiella pneumoniae. Indian J Med Microbiol 37(2):210–218. https://doi.org/10.4103/ijmm.IJMM_19_157
Russo T, Olson R, Macdonald U, Metzger D, Maltese L, Drake E et al (2014) Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 82(6):2356–2367. https://doi.org/10.1128/iai.01667-13
Shakib P, Ghafourian S, Zolfaghary M, Hushmandfar R, Ranjbar R, Sadeghifard N (2012) Prevalence of OmpK35 and OmpK36 porin expression in beta-lactamase and non-betalactamase- producing Klebsiella pneumoniae. Biologics 6:1–4. https://doi.org/10.2147/btt.S27582
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH et al (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain ty**. J Clin Microbiol 33(9):2233–2239. https://doi.org/10.1128/jcm.33.9.2233-2239.1995
Tian D, Pan F, Wang C, Sun Y, Zhang H (2018) Resistance phenotype and clinical molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae among pediatric patients in Shanghai. Infect Drug Resist 11:1935–1943. https://doi.org/10.2147/idr.S175584
Tian Y, Zhang Q, Wen L, Chen J (2021) Combined effect of polymyxin B and tigecycline to overcome heteroresistance in carbapenem-resistant Klebsiella pneumoniae. Microbiol Spectr 9(2):e0015221. https://doi.org/10.1128/Spectrum.00152-21
Turton J, Payne Z, Micah K, Turton J (2018) Capsular type K54, clonal group 29 and virulence plasmids: an analysis of K54 and non-K54 closely related isolates of Klebsiella pneumoniae. Epidemiol Infect 146(14):1813–1823. https://doi.org/10.1017/s0950268818001826
Walsh T, Weeks J, Livermore D, Toleman M (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11(5):355–362. https://doi.org/10.1016/s1473-3099(11)70059-7
Wang Q, Wang X, Wang J, Ouyang P, ** C, Wang R et al (2018) Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis 67(suppl_2):S196-s205. https://doi.org/10.1093/cid/ciy660
Yan Z, Zhou Y, Du M, Bai Y, Liu B, Gong M et al (2019) Prospective investigation of carbapenem-resistant Klebsiella pneumonia transmission among the staff, environment and patients in five major intensive care units. Bei**g J Hosp Infect 101(2):150–157. https://doi.org/10.1016/j.jhin.2018.11.019
Yang X, Dong N, Liu X, Yang C, Ye L, Chan EW et al (2021) Co-conjugation of virulence plasmid and KPC plasmid in a clinical Klebsiella pneumoniae strain. Front Microbiol 12:739461. https://doi.org/10.3389/fmicb.2021.739461
Yu W, Ko W, Cheng K, Lee H, Ke D, Lee C et al (2006) Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 42(10):1351–1358. https://doi.org/10.1086/503420
Yu W, Fung C, Ko W, Cheng K, Lee C, Chuang Y (2007) Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 195(8):1235–1236. https://doi.org/10.1086/512686. (author reply 1236)
Yu W, Ko W, Cheng K, Lee C, Lai C, Chuang Y (2008) Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis 62(1):1–6. https://doi.org/10.1016/j.diagmicrobio.2008.04.007
Zeng L, Yang C, Zhang J, Hu K, Zou J, Li J et al (2021) An outbreak of carbapenem-resistant Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Chongqing, China. Front Cell Infect Microbiol 11:656070. https://doi.org/10.3389/fcimb.2021.656070
Zhan L, Wang S, Guo Y, ** Y, Duan J, Hao Z et al (2017) Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a tertiary hospital in China. Front Cell Infect Microbiol 7:182. https://doi.org/10.3389/fcimb.2017.00182
Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y (2018) Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol 9:289. https://doi.org/10.3389/fmicb.2018.00289
Zhang X, Li F, Cui S, Mao L, Li X, Awan F et al (2020) Prevalence and distribution characteristics of bla(KPC-2) and bla(NDM-1) genes in Klebsiella pneumoniae. Infect Drug Resist 13:2901–2910. https://doi.org/10.2147/idr.S253631
Zhang Y, ** L, Ouyang P, Wang Q, Wang R, Wang J et al (2020) Evolution of hypervirulence in carbapenem-resistant Klebsiella pneumoniae in China: a multicentre, molecular epidemiological analysis. J Antimicrob Chemother 75(2):327–336. https://doi.org/10.1093/jac/dkz446
Zhao Y, Liao Y, Zhang N, Liu S, Zhang J, Hu X et al (2021) Four types of ST11 novel mutations from increasing carbapenem-resistant Klebsiella pneumoniae in Guangdong, 2016–2020. Front Microbiol 12:702941. https://doi.org/10.3389/fmicb.2021.702941
Zhong X, Xu H, Chen D, Zhou H, Hu X, Cheng G (2014) First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS One 9(12):e115185. https://doi.org/10.1371/journal.pone.0115185
Acknowledgements
We sincerely thank all the participants who took part in this study.
Funding
This work was supported by the Science and Technology Program of Hebei Province (No. 20201397), Shijiazhuang Natural Science Foundation (No. 201460523A-2), and the S&T Program of Hebei No. 203777100D.
Author information
Authors and Affiliations
Contributions
Y. G. and Y. L. performed the research. F. L. and Y. G. analyzed the data. Y. L. collected the samples and isolated the strains. W. G. and B. X. provided reagents and materials. Y. Z. and X. W. contributed to the MLST analysis and antimicrobial resistance test. F. L., Y. G., and Y. L. performed the detection of the CRKP serotype, virulence gene, resistant gene, and PFGE. F. L. wrote the first draft of the manuscript. N. S. reviewed and revised this article. All authors have approved to submit the manuscript to your esteemed journal.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study was approved by the Ethics Committee of Shijiazhuang People’s Hospital. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, Y., Liu, F., Zhang, Y. et al. Virulence, antimicrobial resistance, and molecular characteristics of carbapenem-resistant Klebsiella pneumoniae in a hospital in Shijiazhuang City from China. Int Microbiol 26, 1073–1085 (2023). https://doi.org/10.1007/s10123-023-00357-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-023-00357-x