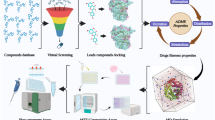
Abstract
Background
Sirtuins (SIRTs) are NAD+-dependent deacetylases that play various roles in numerous pathophysiological processes, holding promise as therapeutic targets worthy of further investigation. Among them, the SIRT2 subtype is closely associated with tumorigenesis and malignancies. Dysregulation of SIRT2 activation can regulate the expression levels of related genes in cancer cells, leading to tumor occurrence and metastasis.
Methods
In this study, we used computer simulations to screen for novel SIRT2 inhibitors from the FDA database, based on which 10 compounds with high docking scores and good interactions were selected for in vitro anti-pancreatic cancer metastasis testing and enzyme binding inhibition experiments. The results showed that fluvastatin sodium may possess inhibitory activity against SIRT2. Subsequently, fluvastatin sodium was subjected to molecular docking experiments with various SIRT isoforms, and the combined results from Western blotting experiments indicated its potential as a SIRT2 inhibitor. Next, molecular docking, molecular dynamics (MD) simulations, and binding free energy calculations were performed, revealing the binding mode of fluvastatin sodium at the SIRT2 active site, further validating the stability and interaction of the ligand–protein complex under physiological conditions.
Results
Overall, this study provides a systematic virtual screening workflow for the discovery of SIRT2 activity inhibitors, identifies the potential inhibitory effect of fluvastatin sodium as a lead compound on SIRT2, and opens up a new direction for develo** highly active and selectively targeted SIRT2 inhibitors.








Similar content being viewed by others
Data availability
Available on request.
References
Kang Z, Wang C, Tong Y et al (2021) Novel nonsecosteroidal vitamin D receptor modulator combined with gemcitabine enhances pancreatic cancer therapy through remodeling of the tumor microenvironment. J Med Chem 64:629–643. https://doi.org/10.1021/acs.jmedchem.0c01197
Wang S-P, Li Y, Huang S-H et al (2021) Discovery of potent and novel dual PARP/BRD4 inhibitors for efficient treatment of pancreatic cancer. J Med Chem 64:17413–17435. https://doi.org/10.1021/acs.jmedchem.1c01535
Ansari D, Tingstedt B, Andersson B et al (2016) Pancreatic cancer: yesterday, today and tomorrow. Future Oncol 12:1929–1946. https://doi.org/10.2217/fon-2016-0010
Hu Y, Guo M (2020) Synthetic lethality strategies: beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci 111:3111–3121. https://doi.org/10.1111/cas.14565
Zhou P, Li B, Liu F et al (2017) The epithelial to mesenchymal transition (EMT) and cancer stem cells: implication for treatment resistance in pancreatic cancer. Mol Cancer 16:52. https://doi.org/10.1186/s12943-017-0624-9
Khalaf N, El-Serag HB, Abrams HR, Thrift AP (2021) Burden of pancreatic cancer: from epidemiology to practice. Clin Gastroenterol Hepatol 19:876–884. https://doi.org/10.1016/j.cgh.2020.02.054
Panchal K, Sahoo RK, Gupta U, Chaurasiya A (2021) Role of targeted immunotherapy for pancreatic ductal adenocarcinoma (PDAC) treatment: an overview. Int Immunopharmacol 95:107508. https://doi.org/10.1016/j.intimp.2021.107508
Bhardwaj VK, Purohit R (2020) A new insight into protein-protein interactions and the effect of conformational alterations in PCNA. Int J Biol Macromol 148:999–1009. https://doi.org/10.1016/j.ijbiomac.2020.01.212
Kumar S, Sinha K, Sharma R et al (2019) Phloretin and phloridzin improve insulin sensitivity and enhance glucose uptake by subverting PPARγ/Cdk5 interaction in differentiated adipocytes. Exp Cell Res 383:111480. https://doi.org/10.1016/j.yexcr.2019.06.025
Palmirotta R, Cives M, Della-Morte D et al (2016) Sirtuins and cancer: role in the epithelial-mesenchymal transition. Oxid Med Cell Longev 2016:3031459. https://doi.org/10.1155/2016/3031459
Bhardwaj V, Purohit R (2020) Computational investigation on effect of mutations in PCNA resulting in structural perturbations and inhibition of mismatch repair pathway. J Biomol Struct Dyn 38:1963–1974. https://doi.org/10.1080/07391102.2019.1621210
Wang Y, Yang J, Hong T et al (2019) SIRT2: Controversy and multiple roles in disease and physiology. Ageing Res Rev 55:100961. https://doi.org/10.1016/j.arr.2019.100961
Kaya SG, Eren G (2024) Selective inhibition of SIRT2: a disputable therapeutic approach in cancer therapy. Bioorganic Chem 143:107038. https://doi.org/10.1016/j.bioorg.2023.107038
Hu F, Sun X, Li G et al (2018) Inhibition of SIRT2 limits tumour angiogenesis via inactivation of the STAT3/VEGFA signalling pathway. Cell Death Dis 10:9. https://doi.org/10.1038/s41419-018-1260-z
Li Y, Zhang M, Dorfman RG et al (2018) SIRT2 promotes the migration and invasion of gastric cancer through RAS/ERK/JNK/MMP-9 pathway by increasing PEPCK1-related metabolism. Neoplasia 20:745–756. https://doi.org/10.1016/j.neo.2018.03.008
Gao C-X, Chen B, **e H-K, et al. (2019) Immunohistochemistry and clinical value of sirtuin 2 in non-metastasized non-small cell lung cancer. J Thorac Dis 11 9 J Thorac Dis
Wei R, He D, Zhang X (2018) Role of SIRT2 in regulation of stemness of cancer stem-like cells in renal cell carcinoma. Cell Physiol Biochem 49:2348–2357. https://doi.org/10.1159/000493835
Chen J, Chan AWH, To K-F et al (2013) SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3β/β-catenin signaling. Hepatology 57:2287–2298. https://doi.org/10.1002/hep.26278
Rajasekaran R, George Priya Doss C, Sudandiradoss C et al (2008) Effect of deleterious nsSNP on the HER2 receptor based on stability and binding affinity with herceptin: a computational approach. C R Biol 331:409–417. https://doi.org/10.1016/j.crvi.2008.03.004
Kamaraj B, Purohit R (2016) Mutational analysis on membrane associated transporter protein (MATP) and their structural consequences in oculocutaeous albinism type 4 (OCA4)—a molecular dynamics approach. J Cell Biochem 117:2608–2619. https://doi.org/10.1002/jcb.25555
Singh R, Bhardwaj V, Purohit R (2021) Identification of a novel binding mechanism of quinoline based molecules with lactate dehydrogenase of Plasmodium falciparum. J Biomol Struct Dyn 39:348–356. https://doi.org/10.1080/07391102.2020.1711809
Zhao D, Zou S-W, Liu Y et al (2013) Lysine-5 acetylation negatively regulates lactate dehydrogenase a and is decreased in pancreatic cancer. Cancer Cell 23:464–476. https://doi.org/10.1016/j.ccr.2013.02.005
Liu PY, Xu N, Malyukova A et al (2013) The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ 20:503–514. https://doi.org/10.1038/cdd.2012.147
Grozinger CM, Chao ED, Blackwell HE et al (2001) Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening*. J Biol Chem 276:38837–38843. https://doi.org/10.1074/jbc.M106779200
Taylor DM, Balabadra U, **ang Z et al (2011) A brain-permeable small molecule reduces neuronal cholesterol by inhibiting activity of sirtuin 2 deacetylase. ACS Chem Biol 6:540–546. https://doi.org/10.1021/cb100376q
**g H, Hu J, He B et al (2016) A SIRT2-selective inhibitor promotes c-Myc oncoprotein degradation and exhibits broad anticancer activity. Cancer Cell 29:297–310. https://doi.org/10.1016/j.ccell.2016.02.007
Spiegelman NA, Hong JY, Hu J et al (2019) A small-molecule SIRT2 inhibitor that promotes K-Ras4a lysine fatty-acylation. ChemMedChem 14:744–748. https://doi.org/10.1002/cmdc.201800715
Lin X, Li X, Lin X (2020) A review on applications of computational methods in drug screening and design. Molecules 25:. https://doi.org/10.3390/molecules25061375
Hassan Baig M, Ahmad K, Roy S, et al. (2015) Computer aided drug design: success and limitations. Curr Pharm Des
Song CM, Lim SJ, Tong JC (2009) Recent advances in computer-aided drug design. Brief Bioinform 10:579–591. https://doi.org/10.1093/bib/bbp023
Macalino SJY, Gosu V, Hong S, Choi S (2015) Role of computer-aided drug design in modern drug discovery. Arch Pharm Res 38:1686–1701. https://doi.org/10.1007/s12272-015-0640-5
Friesner RA, Murphy RB, Repasky MP et al (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/jm051256o
Wu J, Hu B, Lu S et al (2022) Identification of raloxifene as a novel α-glucosidase inhibitor using a systematic drug repurposing approach in combination with cross molecular docking-based virtual screening and experimental verification. Carbohydr Res 511:108478. https://doi.org/10.1016/j.carres.2021.108478
Halgren TA, Murphy RB, Friesner RA et al (2004) Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem 47:1750–1759. https://doi.org/10.1021/jm030644s
Roos K, Wu C, Damm W et al (2019) OPLS3e: extending force field coverage for drug-like small molecules. J Chem Theory Comput 15:1863–1874. https://doi.org/10.1021/acs.jctc.8b01026
Yadava U, Shukla BK, Roychoudhury M, Kumar D (2015) Pyrazolo[3,4-d]pyrimidines as novel inhibitors of O-acetyl-l-serine sulfhydrylase of Entamoeba histolytica: an in silico study. J Mol Model 21:96. https://doi.org/10.1007/s00894-015-2631-3
Ylilauri M, Pentikäinen OT (2013) MMGBSA as a tool to understand the binding affinities of filamin–peptide interactions. J Chem Inf Model 53:2626–2633. https://doi.org/10.1021/ci4002475
You W, Steegborn C (2020) Structural basis for activation of human sirtuin 6 by fluvastatin. ACS Med Chem Lett 11:2285–2289. https://doi.org/10.1021/acsmedchemlett.0c00407
Funding
The research was financially supported by the Natural Science Foundation of Liaoning Province (2022-MS-247), the Career Development Support Program for Young and Middle-aged Teachers at Shenyang Pharmaceutical University (ZQN2021001), the General Project of Liaoning Province Education Department (JYTMS20231359), and the National College Students Innovation and Entrepreneurship Training Program (202210163034).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Study conception and design: ** Yang, Chao Ma. Acquisition of data: ** Yang, Hanxun Wang, Enlong Ma. Analysis and interpretation of data: ** Yang, Hanxun Wang, Jiale Liu. Drafting of the manuscript: ** Yang. Critical revision: Yanchun Li, **nxin **. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This article does not contain any studies involving animals or human participants performed by any of the authors.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Wang, H., Liu, J. et al. Screening approach by a combination of computational and in vitro experiments: identification of fluvastatin sodium as a potential SIRT2 inhibitor. J Mol Model 30, 188 (2024). https://doi.org/10.1007/s00894-024-05988-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05988-z