Abstract
Context
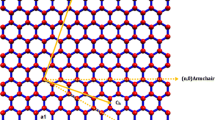
Graphene based nano sensors have huge potential in an era of sensor technology. The objective of this study is to create a sensor by investigating the vibration responses of cantilever and bridged boundary conditioned single layer graphene sheets (SLGS) with various attached microorganisms on the tip and at the centre of the sheet. The Parvoviridae, Flaviviridae, and Polyomaviridae biological substances have been comprehensively investigated here. For the Parvoviridae, Polyomaviridae, and Flaviviridae categories of targeted microbes, the sizes are 21nm, 40nm, and 45nm, respectively. The Parvoviridae family has a maximum frequency of 1.87x107 Hz with a cantilever condition and a mass of 4.2441 Zg, and for a bridged condition, it demonstrates a maximum frequency of 1.23x108 Hz with the same mass on armchair SLG (5 5). The data analysis shows that 3.0041 Zg mass of the Mimivirus has the lowest frequency. It demonstrates explicitly that the rate of frequency decreases as the value of mass increases. When compared to chiral SLG, the armchair single layer graphene sheet performs better. The research indicates that the dynamic properties are significantly influenced by the mass of various biological organisms. The application of this sensor will enable the detection of microorganisms or viruses that can be connected to SLG.
Methods
In this research, the application of Single Layer Graphene (SLG) as a virus sensing device is explored. Atomistic finite element method (AFEM) has been used to carry out the dynamic analysis of SLG. Molecular dynamic analysis and simulations have been performed to see how SLG behaves when employed as sensors for biological entities and when they are exposed to bridged and cantilever boundary conditions. The frequency analysis was performed using ANSYS APDL software. SLG of various chirality has been utilised in the investigation. By altering the applied mass of a biological object, the difference in frequency observed. The idea behind mass detection employing nano biosensors is built on the concept that the stiffness of a biomolecule changes as its mass changes, making the resonant frequency extremely sensitive to that change. A shift in the resonance frequency results from a change in the associated mass on the graphene sheet. The main challenge in mass detection is estimating the variation in resonant frequency driven by the mass of the connected molecule. The SLG-based biosensor has a specific application in the early identification of diseases. The biosensor investigated in this article is novel, whereas the biosensors that are presently on the market operate using the ionization method. The simulations result shows SLG based biosensor's sensitivity considerably faster than an existing one.




















Similar content being viewed by others
References
Li Y, Luo G, Qing Z, Li X, Zou Z, Yang R (2019) Colorimetric aminotriazole assay based on catalase deactivation-dependent longitudinal etching of gold nanorods. Microchim Acta 186(8):1–7
Qing Z, Bai A, Chen L, **ng S, Zou Z, Lei Y, Yang R (2021) An activatable nanoenzyme reactor for coenhanced chemodynamic and starving therapy against tumor hypoxia and antioxidant defense system. CCS Chem 3(5):1217–1230
Qing Z, Luo G, **ng S, Zou Z, Lei Y, Liu J, Yang R (2020) Pt–S bond-mediated nanoflares for high-fidelity intracellular applications by avoiding thiol cleavage. Angew Chem Int Ed 59(33):14044–14048
Sanvicens N, Pastells C, Pascual N, Marco MP (2009) Nanoparticle-based biosensors for detection of pathogenic bacteria. TrAC Trends Anal Chem 28(11):1243–1252
Justino CI, Gomes AR, Freitas AC, Duarte AC, Rocha-Santos TA (2017) Graphene based sensors and biosensors. TrAC Trends Anal Chem 91:53–66
Zhang H, Zhang H, Aldalbahi A, Zuo X, Fan C, Mi X (2017) Fluorescent biosensors enabled by graphene and graphene oxide. Biosens Bioelectron 89:96–106
Zhu C, Du D, Lin Y (2017) Graphene-like 2D nanomaterial-based biointerfaces for biosensing applications. Biosens Bioelectron 89:43–55
Forsyth R, Devadoss A, Guy OJ (2017) Graphene field effect transistors for biomedical applications: current status and future prospects. Diagnostics 7(3):45
Kolev SK, Aleksandrov HA, Atanasov VA, Popov VN, Milenov TI (2019) Interaction of graphene with out-of-plane aromatic hydrocarbons. J Phys Chem C 123(35):21448–21456
Danielson E, Sontakke VA, Porkovich AJ, Wang Z, Kumar P, Ziadi Z, Sowwan M (2020) Graphene based field-effect transistor biosensors functionalized using gas-phase synthesized gold nanoparticles. Sens Actuators B Chem 320:128432
Ahmed A, Rushworth JV, Hirst NA, Millner PA (2014) Biosensors for whole-cell bacterial detection. Clin Microbiol Rev 27(3):631–646
Xu Y, **e X, Duan Y, Wang L, Cheng Z, Cheng J (2016) A review of impedance measurements of whole cells. Biosens Bioelectron 77:824–836
Ajayan PM, Charlier JC, Rinzler AG (1999) Carbon nanotubes: from macromolecules to nanotechnology. Proc Natl Acad Sci 96(25):14199–14200
Chowdhury R, Adhikari S (2010) Boron-nitride nanotubes as zeptogram-scale bionanosensors: theoretical investigations. IEEE Trans Nanotechnol 10(4):659–667
Chowdhury R, Adhikari S, Mitchell J (2009) Vibrating carbon nanotube-based bio-sensors. Physica E 42(2):104–109
Georgantzinos SK, Anifantis NK (2010) Carbon nanotube-based resonant nanomechanical sensors: a computational investigation of their behavior. Physica E 42(5):1795–1801
Elishakoff I, Versaci C, Maugeri N, Muscolino G (2011) Clamped-free single-walled carbon nanotube-based mass sensor treated as Bernoulli–Euler beam. J Nanotechnol Eng Med 2(2)
Tounsi A, Semmah A, Bousahla AA (2013) Thermal buckling behavior of nanobeams using an efficient higher-order nonlocal beam theory. J Nanomech Micromech 3(3):37–42
Iijima S, Brabec C, Maiti A, Bernholc J (1996) Structural flexibility of carbon nanotubes. J Chem Phys 104(5):2089–2092
Iijima S (1991) Helical microtubules of graphitic carbon. Nature 354(6348):56–58
Li C, Chou TW (2004) Vibrational behaviors of multiwalled-carbon-nanotube-based nanomechanical resonators. Appl Phys Lett 84(1):121–123
Li C, Chou TW (2004) Mass detection using carbon nanotube-based nanomechanical resonators. Appl Phys Lett 84(25):5246–5248
Hernández R, Vallés C, Benito AM, Maser WK, Rius FX, Riu J (2014) Graphene-based potentiometric biosensor for the immediate detection of living bacteria. Biosens Bioelectron 54:553–557
Joshi AY, Patel AM (2017) Sensing the presence and amount of microbes using double walled carbon nanotubes. Advancing Medicine through Nanotechnology and Nanomechanics Applications. IGI Global, pp 78–117
Bhardwaj SK, Bhardwaj N, Kumar V, Bhatt D, Azzouz A, Bhaumik J, Kim K-H, Deep A (2021) Recent progress in nanomaterial-based sensing of airborne viral and bacterial pathogens. Environ Int 146:106183
Moens U, Calvignac-Spencer S, Lauber C, Ramqvist T, Feltkamp MC, Daugherty MD, Ehlers B (2017) ICTV virus taxonomy profile: Polyomaviridae. J Gen Virol 98(6):1159–1160
Harprecht C (2020) Structural analysis of JC polyomavirus inhibitors and receptors (Doctoral dissertation). Universität Tübingen
Taxonomy, V. (2020). Release. International Committee on Taxonomy of Viruses (ICTV). March 2019. Archived from the original on 2018-03-04. Retrieved 2020-01-24
Gorbalenya AE, Snijder EJ, Spaan WJ (2004) Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol 78(15):7863–7866
Minutilli E, Mulè A (2016) Merkel cell polyomavirus and cutaneous Merkel cell carcinoma. Future Sci OA 2(4):FSO155
Gossai A, Waterboer T, Nelson HH, Michel A, Willhauck-Fleckenstein M, Farzan SF, Hoen AG et al (2016) Seroepidemiology of human polyomaviruses in a US population. Am J Epidemiol 183(1):61–69
Knowles WA, Pipkin P, Andrews N, Vyse A, Minor P, Brown DW, Miller E (2003) Population-based study of antibody to the human polyomaviruses BKV and JCV and the simian polyomavirus SV40. J Med Virol 71(1):115–123
Jamboti JS (2016) BK virus nephropathy in renal transplant recipients. Nephrology 21(8):647–654
Kuppachi S, Kaur D, Holanda DG, Thomas CP (2016) BK polyoma virus infection and renal disease in non-renal solid organ transplantation. Clin Kidney J 9(2):310–318
Adang L, Berger J (2015) Progressive multifocal leukoencephalopathy. F1000Research 4
Feng H, Shuda M, Chang Y, Moore PS (2008) Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319(5866):1096–1100
Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S et al (2017) ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98(1):2–3
Scheel TK, Simmonds P, Kapoor A (2015) Surveying the global virome: identification and characterization of HCV-related animal hepaciviruses. Antivir Res 115:83–93
Valles SM, Chen Y, Firth AE, Guérin DMA, Hashimoto Y, Herrero S, De Miranda JR, Ryabov E, ICTV Report Consortium (2017) ICTV virus taxonomy profile: Iflaviridae. J Gen Virol 98(4):527–528
Rosario K, Breitbart M, Harrach B, Segalés J, Delwart E, Biagini P, Varsani A (2017) Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol 162(5):1447–1463
Li C, Chou TW (2003) A structural mechanics approach for the analysis of carbon nanotubes. Int J Solids Struct 40(10):2487–2499
Stuart SJ, Tutein AB, Harrison JA (2000) A reactive potential for hydrocarbons with intermolecular interactions. J Chem Phys 112(14):6472–6486
Blagov EV, Klimchitskaya GL, Mostepanenko VM (2007) van der Waals interaction between a microparticle and a single-walled carbon nanotube. Phys Rev B 75(23):235413
Zhang YY, Wang CM, Tan VBC (2009) Assessment of Timoshenko beam models for vibrational behavior of single-walled carbon nanotubes using molecular dynamics. Adv Appl Math Mech 1(1):89–106
Lee HL, Chang WJ (2009) A closed-form solution for critical buckling temperature of a single-walled carbon nanotube. Physica E 41(8):1492–1494
Natsuki T (2015) Theoretical analysis of vibration frequency of graphene sheets used as nanomechanical mass sensor. Electronics 4(4):723–738
Makwana M, Patel AM, Oza AD, Prakash C, Gupta LR, Vatin NI, Dixit S (2022) Effect of mass on the dynamic characteristics of single-and double-layered graphene-based nano resonators. Materials 15(16):5551
Makwana MV, Patel AM (2022) Molecular dynamic analysis of pristine single layered graphene for mass sensor. Materials Today: Proceedings
Makwana MV, Patel AM (2022) Recent applications and synthesis techniques of graphene. Micro Nanosyst 14(4):287–303
Makwana M, Patel AM (2023) Nanoresonator vibrational behaviour analysis of single-and double-layer graphene with atomic vacancy and pinhole defects. J Mol Model 29(5):149
Elishakoff I, Challamel N, Soret C, Bekel Y, Gomez T (2013) Virus sensor based on single-walled carbon nanotube: improved theory incorporating surface effects. Philos Trans Royal Soc A: Math, Phys Eng Sci 371(1993):20120424
Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P (2010) ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 39(Database issue):D576–D582
Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P (2011) ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 39(suppl_1):D576–D582. https://doi.org/10.1093/nar/gkq901
Bofill-Mas, S. (2016). Polyomavirus. In: J.B. Rose and B. Jiménez-Cisneros, (eds) Water and sanitation for the 21st century: health and microbiological aspects of excreta and wastewater management (Global Water Pathogen Project). (J.S Meschke, and R. Girones (eds), Part 3: specific excreted pathogens: environmental and epidemiology aspects - Section 1: Viruses), Michigan State University, E. Lansing, MI, UNESCO
Simmonds P, Becher P, Bukh J, Gould EA, Meyers G, Monath T, Muerhoff S, Pletnev A, Rico-Hesse R, Smith DB, Stapleton JT, Ictv Report Consortium (2017) ICTV virus taxonomy profile: Flaviviridae. J Gen Virol 98(1):2–3. https://doi.org/10.1099/jgv.0.000672 PMID: 28218572; PMCID: PMC5370391
Mehranfar A, Khavani M, Izadyar M (2022) A molecular dynamic study on the ability of phosphorene for designing new sensor for SARS-CoV-2 detection. J Mol Liq 345:117852
Verma N, Badhe Y, Gupta R, Maparu AK, Rai B (2022) Peptide mediated colorimetric detection of SARS-CoV-2 using gold nanoparticles: a molecular dynamics simulation study. J Mol Model 28(7):202
Gholami R, Solimannejad M (2022) Potential of B24N24 nanocluster for sensing and delivering aloe-emodin anticancer drug: A DFT study. J Mol Struct 1270:133968
Mayo MA, Pringle CR (1998) Virus taxonomy-1997. J Gen Virol 79(4):649–657
Murphy FA, Fauquet CM, Bishop DH, Ghabrial SA, Jarvis AW, Martelli GP, Summers MD (2012) Virus taxonomy: classification and nomenclature of viruses (Vol. 10). Springer Science & Business Media
EA G, Solomon T (2008) Pathogenic flaviviruses. Lancet 371(9611):500–509
Qiu Y, Zhang Y, Ademiloye AS, Wu Z (2020) Molecular dynamics simulations of single-layer and rotated double-layer graphene sheets under a high velocity impact by fullerene. Comput Mater Sci 182:109798
Chaudhary R, Mudimela PR (2020) 3D modeling of graphene oxide based nanoelectromechanical capacitive switch. Microsyst Technol 26(9):2931–2937
Chaudhary R, Mudimela PR (2020) Pull-in response and eigen frequency analysis of graphene oxide-based NEMS switch. Mater Today: Proc 28:196–200
Chaudhary R, Jhanwar P, Mudimela PR (2023) Finite element analysis of graphene oxide hinge structure-based RF NEM switch. IETE J Res 69(2):967–974
Kuila T, Bose S, Khanra P, Mishra AK, Kim NH, Lee JH (2011) Recent advances in graphene-based biosensors. Biosens Bioelectron 26(12):4637–4648
Huang Y, Dong X, Liu Y, Li LJ, Chen P (2011) Graphene-based biosensors for detection of bacteria and their metabolic activities. J Mater Chem 21(33):12358–12362
Shearer CJ, Slattery AD, Stapleton AJ, Shapter JG, Gibson CT (2016) Accurate thickness measurement of graphene. Nanotechnology 27(12):125704
Acknowledgements
The authors acknowledge the CVM University, Vallabh Vidyanagar, Gujarat, India.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by [Manisha Makwana] and [Dr. Ajay M Patel]. The first draft of the manuscript was written by [Manisha Makwana], and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Conflict of interest
The authors declare no competing interests.
Human and animal rights
Our manuscript does not report on or involve the use of any animal or human data or tissue.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Makwana, M., Patel, A.M. Identification of microbes using single-layer graphene-based nano biosensors. J Mol Model 29, 382 (2023). https://doi.org/10.1007/s00894-023-05748-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-023-05748-5