Abstract
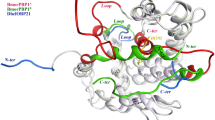
The odorant binding protein of Culex quinquefasciatus (CquiOBP1), expressed on the insect antenna, is crucial for the investigation of trap** baited with oviposition semi-chemicals and controlling mosquito populations. The acidic titratable residues pKa prediction and the ligand binding poses investigation in two systems (pH 7 and pH 5) are studied by constant pH molecular dynamics (CpHMD) and molecular docking methods. Research results reveal that the change of the protonation states would disrupt some important H-bonds, such as Asp 66-Asp 70, Glu 105-Asn 102, etc. The cleavage of these H-bonds leads to the movement of the relative position of hydrophobic tunnel, N- and C- termini loops and pH-sensing triad (His23-Tyr54-Val125) in acid solution. Ligand MOP has lower affinity and shows different binding poses to protein CquiOBP1 at pH 5. This ligand may be released from another tunnel between helices α3 and α4 in acidic environment. However, it would bind to the protein with high affinity in neutral environment. This work could provide more penetrating understanding of the pH-induced ligand-releasing mechanism.







Similar content being viewed by others
References
Benton R (2006) On the origin of smell: odorant receptors in insects. Cell Mol Life Sci 63(14):1579–1585
Pelosi P, Calvello M, Ban L (2005) Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem Senses 30(suppl 1):i291–i292
Pelosi P, Zhou JJ, Ban L, Calvello M (2006) Soluble proteins in insect chemical communication. Cell Mol Life Sci 63(14):1658–1676
Pelosi P, Maida R (1995) Odorant-binding proteins in insects. Comp Biochem Physiol B 111(3):503–514
Zhou JJ (2010) Odorant-binding proteins in insects. Vitam Horm 83:241–272
Chandre F, Darriet F, Darder M, Cuany A, Doannio J, Pasteur N, Guillet P (1998) Pyrethroid resistance in Culex quinquefasciatusfrom West Africa. Med Vet Entomol 12(4):359–366
Barbosa RMR, Furtado A, Regis L, Leal WS (2010) Evaluation of an oviposition stimulating kairomone for the yellow fever mosquito, Aedes aegypti, in Recife, Brazil. J Vector Ecol 35(1):204–207
Leal WS, Barbosa R, Xu W, Ishida Y, Syed Z, Latte N, Chen AM, Morgan TI, Cornel AJ, Furtado A (2008) Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes. PLoS One 3(8):e3045
Horst R, Damberger F, Luginbühl P, Güntert P, Peng G, Nikonova L, Leal WS, Wüthrich K (2001) NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc Natl Acad Sci U S A 98(25):14374
Leal WS, Chen AM, Erickson ML (2005) Selective and pH-dependent binding of a moth pheromone to a pheromone-binding protein. J Chem Ecol 31(10):2493–2499
Gräter F, de Groot BL, Jiang H, Grubmüller H (2006) Ligand-release pathways in the pheromone-binding protein of Bombyx mori. Structure 14(10):1567–1576
Leite NR, Krogh R, Xu W, Ishida Y, Iulek J, Leal WS, Oliva G (2009) Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive “lid”. PLoS One 4(11):e8006
Mao Y, Xu X, Xu W, Ishida Y, Leal WS, Ames JB, Clardy J (2010) Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc Natl Acad Sci U S A 107(44):19102
Mertz JE, Pettitt BM (1994) Molecular dynamics at a constant pH. Int J High Perform Comput Appl 8(1):47
Baptista AM, Martel PJ, Petersen SB (1997) Simulation of protein conformational freedom as a function of pH: constant pH molecular dynamics using implicit titration. Proteins 27(4):523–544
Börjesson U, Hünenberger PH (2001) Explicit-solvent molecular dynamics simulation at constant pH: methodology and application to small amines. J Chem Phys 114:9706
Börjesson U, Hünenberger PH (2004) pH-dependent stability of a decalysine α-helix studied by explicit-solvent molecular dynamics simulations at constant pH. J Phys Chem B 108(35):13551–13559
Khandogin J, Brooks CL (2005) Constant pH molecular dynamics with proton tautomerism. Biophys J 89(1):141–157
Lee MS (2004) Constant-pH molecular dynamics using continuous titration coordinates. Proteins 56:738–752
Khandogin J, Brooks CL (2007) Linking folding with aggregation in Alzheimer’s α-amyloid peptides. Proc Natl Acad Sci U S A 104(43):16880
Khandogin J, Chen J, Brooks CL (2006) Exploring atomistic details of pH-dependent peptide folding. Proc Natl Acad Sci U S A 103(49):18546
Mongan J, Case DA, McCammon JA (2004) Constant pH molecular dynamics in generalized born implicit solvent. J Comput Chem 25(16):2038–2048
Baptista AM, Teixeira VH, Soares CM (2002) Constant-pH molecular dynamics using stochastic titration. J Chem Phys 117:4184
Machuqueiro M, Baptista AM (2006) Constant-pH molecular dynamics with ionic strength effects: protonation-conformation coupling in decalysine. J Phys Chem B 110(6):2927–2933
Machuqueiro M, Baptista AM (2009) Molecular dynamics at constant pH and reduction potential: application to cytochrome c 3. J Am Chem Soc 131(35):12586–12594
Bürgi R, Kollman PA, van Gunsteren WF (2002) Simulating proteins at constant pH: an approach combining molecular dynamics and Monte Carlo simulation. Protein Struct Funct Bioinforma 47(4):469–480
Dlugosz M, Antosiewicz J (2005) Effects of solute-solvent proton exchange on polypeptide chain dynamics: a constant-pH molecular dynamics study. J Phys Chem B 109(28):13777–13784
Walczak AM, Antosiewicz JM (2002) Langevin dynamics of proteins at constant pH. Phys Rev E 66(5):051911
Williams SL, De Oliveira CAF, McCammon JA (2010) Coupling constant pH molecular dynamics with accelerated molecular dynamics. J Chem Theory Comput 6(2):560–568
Meng Y, Roitberg AE (2010) Constant pH replica exchange molecular dynamics in biomolecules using a discrete protonation model. J Chem Theory Comput 6(4):1401–1412
Bashford D, Case DA (2000) Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem 51(1):129–152
Onufriev A, Bashford D, David A (2000) Modification of the generalized born model suitable for macromolecules. J Phys Chem B 104(15):3712–3720
Case DA, Darden T, Cheatham III TE, Simmerling C, Wang J, Duke RE, Luo R, Crowley M, Walker R, Zhang W (2008) Amber 10 users manual. University of California
Onufriev A, Case DA, Bashford D (2002) Effective Born radii in the generalized born approximation: the importance of being perfect. J Comput Chem 23(14):1297–1304
Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55(2):383–394
Berendsen HJC, Postma JPM, Van Gunsteren WF, DiNola A, Haak J (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Sham YY, Muegge I, Warshel A (1998) The effect of protein relaxation on charge-charge interactions and dielectric constants of proteins. Biophys J 74(4):1744–1753
Simonson T, Archontis G, Karplus M (1999) A Poisson-Boltzmann study of charge insertion in an enzyme active site: the effect of dielectric relaxation. J Phys Chem B 103(29):6142–6156
Warshel A, Aqvist J (1991) Electrostatic energy and macromolecular function. Annu Rev Biophys Biophys Chem 20(1):267–298
Lautenschlager C, Leal WS, Clardy J (2005) Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem Biophys Res Commun 335(4):1044–1050
Xu W, Leal WS (2008) Molecular switches for pheromone release from a moth pheromone-binding protein. Biochem Biophys Res Commun 372(4):559–564
Acknowledgments
This work is supported by Natural Science Foundation of China, Specialized Research Fund for the Doctoral Program of Higher Education, and Specialized Fund for the Basic Research of Jilin University (Grant Nos. 20903045, 20573042, 20070183046, and 200810018).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 283 kb)
Rights and permissions
About this article
Cite this article
Chu, WT., Zhang, JL., Zheng, QC. et al. Constant pH molecular dynamics (CpHMD) and molecular docking studies of CquiOBP1 pH-induced ligand releasing mechanism. J Mol Model 19, 1301–1309 (2013). https://doi.org/10.1007/s00894-012-1680-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1680-0