Abstract
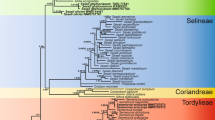
An expanded phylogenetic reconstruction based on the nuclear ribosomal DNA internal transcribed spacer region (nrDNA ITS) in conjunction with a multivariate statistical analysis of morphological characters revealed a new species and the re-acknowledgement of another one in Sclerorhachis (Compositae, Anthemideae). The newly revealed species, Sclerorhachis ferdowsii, has been previously included in the so-called S. platyrachis-complex, but is easily distinguished as an independent species by its rhizomatous root system, sparsely paleate receptacles, coronate and costate achenes, the relatively smaller size of the capitula, and the smaller habit of the whole plant. Additionally, morphological and molecular data corroborated S. paropamisica as a distinct species rather than being conspecific with S. platyrachis. With these newly acknowledged taxa, the number of species in Sclerorhachis is now expanded to eight. A detailed morphological description, an illustration, and distribution maps for S. ferdowsii, along with an identification key for all species of Sclerorhachis, are provided.







Similar content being viewed by others
Data availability
All DNA sequences are publicly available through Genbank. All other data included in the manuscript.
References
Abdesselam R. (2010) Discriminant Analysis on Mixed Predictors. In: Palumbo F., Lauro C., Greenacre M. (eds) Data Analysis and Classification. Studies in Classification, Data Analysis, and Knowledge Organization. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-03739-9_13
Alvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molec Phylogen Evol 29:417–434. https://doi.org/10.1016/S1055-7903(03)00208-2
Bachman S, Moat J, Hill AW, De La Torre J, Scott B (2011) Supporting red list threat assessments with GeoCAT: geospatial conservation assessment tool. ZooKeys 150:117-126. https://doi.org/10.3897/zookeys.150.2109
Capella-Gutierrez S, Kauff F, Gabaldon T (2014) A phylogenomics approach for selecting robust sets of phylogenetic markers. Nucl Acids Res 42:e54–e54. https://doi.org/10.1093/nar/gku071
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Meth 9:772. https://doi.org/10.1038/nmeth.2109
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Franzke A, German D, Al-Shehbaz IA, Mummenhoff K (2009) Arabidopsis family ties: molecular phylogeny and age estimates in Brassicaceae. Taxon 58:425–437. https://doi.org/10.1002/tax.582009
Hassanpour H, Zare H, Ali M et al (2018) Phylogenetic species delimitation unravels a new species in the genus Sclerorhachis (Rech. f.) Rech. f. (Compositae, Anthemideae ). Pl Syst Evol 304:185–203. https://doi.org/10.1007/s00606-017-1461-4
Hulsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Bioinformatics 17:754–755
Iranshahr M (1979) Sclerorhachis rechingeri (Asteraceae-Anthemideae), a new species from N. Khorasan. Pl Syst Evol 132:149–152
IUCN (2021) The IUCN Red List of Threatened Species. Version 2021–3. Available at: https://www.iucnredlist.org. Accessed 6 Jun 2022
Joly S, Starr JR, Lewis WH, Bruneau A (2006) Polyploid and hybrid evolution in roses east of the Rocky Mountains. Amer J Bot 93:412–425
Kovalevskaja, SS (1961) De Pyrethro Mucronato Rgl. et Schmalh. Not Syst Herb Inst Bot Acad Sci Uzbekistan 16: 22-31 (in Russian)
Kovalevskaja, SS (1972) Taxa nova generis Tanacetopsis Kovalevsk. Not Syst Herb Inst Bot Acad Sci Uzbekistan 9: 269-271 (in Russian)
Kovalevskaja SS (1987) Sclerorhachis paropamisica (Kurbanov) Koval-ovsk. Novosti Sist Vyssh Rast 24:168 (in Russian)
Krascheninnikov HM (1946) Compositarum species novae. Bot Mater Gerb Bot Inst Komarova Akad Nauk SSSR 9:152─184 (in Russian)
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18
Memariani F, Akhani H, Joharchi MR (2016a) Endemic plants of Khorassan-Kopet Dagh floristic province in Irano-Turanian region: diversity, distribution patterns and conservation status. Phytotaxa 249:31–72. https://doi.org/10.11646/phytotaxa.249.1.5
Memariani F, Zarrinpour V, Akhani H (2016b) A review of plant diversity, vegetation, and phytogeography of the Khorassan-Kopet Dagh floristic province in the Irano-Turanian region (northeastern Iran–southern Turkmenistan). Phytotaxa 249:8–30. https://doi.org/10.11646/phytotaxa.249.1.4
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Molec Biol Evol 30:1188–1195. https://doi.org/10.1093/molbev/mst024
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Molec Biol Evol 37:1530–1534. https://doi.org/10.1093/molbev/msaa131
Moazzeni H, Zarre S, Pfeil BE et al (2014) Phylogenetic perspectives on diversification and character evolution in the species-rich genus Erysimum (Erysimeae; Brassicaceae) based on a densely sampled ITS approach. Bot J Linn Soc 175:497–522. https://doi.org/10.1111/boj.12184
Mohammadi T, Pirani A, Vaezi J, Moazzeni H (2020) A contribution to ethnobotany and review of phytochemistry and biological activities of the Iranian local endemic species Sclerorhachis leptoclada Rech. f. Ethnobot Res Appl 20:1–18
Mozaffarian V (2009) Flora of Iran, No. 59: Compositae: Anthemideae & Echinopeae tribes. Forest and rangeland research institute, Tehran
Oberprieler C (2022) The Wettstein tesseract: a tool for conceptualising species-rank decisions and illustrating speciation trajectories. Taxon. https://doi.org/10.1002/tax.12825
Oberprieler C, Himmelreich S, Vogt R (2007) A new subtribal classification of the tribe Anthemideae (Compositae ). Willdenowia 37:89–114. https://doi.org/10.3372/wi.37.37104
Oberprieler A, Oberprieler C, Töpfer A et al (2022) An updated subtribal classification of Compositae tribe Anthemideae based on extended phylogenetic reconstructions. Willdenowia 52:117–149. https://doi.org/10.3372/wi.52.52108
Pagès J (2004) Analyse factorielle de donnees mixtes. Rev Stat Appl 93–111
Poljakov, P (1959) De generibus Cancrinia Kar. et Kir. et Trichanthemis Rgl. et Schm. Bot Mater Gerb Bot Inst Komarova Akad Nauk SSSR 19:367─379 (in Russian)
Rechinger KH (1986) Sclerorhachis. In: Rechinger KH (ed) Flora Iranica. Akademische Druck-und Verlagsanstalt, Graz, pp 45–48
Sang T, Crawford DJ, Stuessy TF (1995) Documentation of reticulate evolution in peonies (Paeonia) using internal transcribed spacer sequences of nuclear ribosomal DNA: implications for biogeography and concerted evolution. Proc Natl Acad Sci USA 92:6813–6817
Tekpinar AD, Kalmer A (2019) Utility of various molecular markers in fungal identification and phylogeny. Nova Hedwigia 109:187–224. https://doi.org/10.1127/nova
Tzvelev N (1961) Cancrinia paropamisica (Krasch.) Tezvel. In: Schischkin B, Bobrov E (eds) Flora USSR, vol. 26. Bishen Singh Mahendra Pal Singh, Koeltz Science Books, Königstein, pp 296–297
Urantowka AD, Kroczak A, Mackiewicz P (2017) The influence of molecular markers and methods on inferring the phylogenetic relationships between the representatives of the Arini (parrots, Psittaciformes), determined on the basis of their complete mitochondrial genomes. BMC Evol Biol 17:1–26. https://doi.org/10.1186/s12862-017-1012-1
Von Wettstein R (1898) Grundzüge der geographisch-morphologischen Methode der Pflanzensystematik. Jena: Fischer. https://doi.org/10.5962/bhl.title.95473
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 18:315–322
Acknowledgements
The authors are grateful to Dr. Farshid Memariani (Ferdowsi University of Mashhad) for his guidance in preparing the conservation status of the species. Khadijeh Motahhari is appreciated for preparing the distribution maps. We are thankful to the following herbaria: B, E, FUMH, IRAN, K, LE, MPH, TARI, M, MSB, P, and W for assisting with the material review and sending type images.
Funding
This study was partly funded by the Ferdowsi University of Mashhad through the research project 48774–2.
Author information
Authors and Affiliations
Contributions
SH: Supervising the study, specimen study, plant collection, laboratory procedures, molecular analyses, and manuscript preparation. HM: Supervising the study, specimen study, plant collection, laboratory procedures, molecular analyses, and manuscript preparation. AS: providing some references, manuscript revision. SSH: laboratory procedures, manuscript revision. AP: molecular analyses, manuscript revision. MRJ: Specimen study, manuscript revision. CO: providing some references, manuscript revision.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Karol Marhold.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hassanpour, S., Moazzeni, H., Sonboli, A. et al. Molecular and morphological data reveal a new species of Sclerorhachis (Compositae, Anthemideae) and the reassessment of another species of the genus. Plant Syst Evol 309, 10 (2023). https://doi.org/10.1007/s00606-022-01840-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00606-022-01840-0