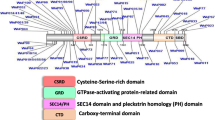
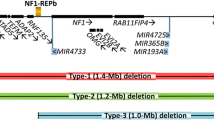
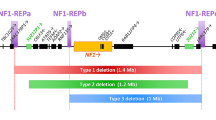
Abstract
Neurofibromatosis type 1 (NF1) is an autosomal dominant disease with complete penetrance but highly variable expressivity. In most patients, Next Generation Sequencing (NGS) technologies allow the identification of a loss-of-function pathogenic variant in the NF1 gene, a negative regulator of the RAS-MAPK pathway. We describe the 5-year diagnosis wandering of a patient with a clear NF1 clinical diagnosis, but no molecular diagnosis using standard molecular technologies. The patient presented with a typical NF1 phenotype but NF1 targeted NGS, NF1 transcript analysis, MLPA, and array comparative genomic hybridization failed to reveal a genetic aberration. After 5 years of unsuccessful investigations, trio WGS finally identified a de novo mosaic (VAF ~ 14%) 24.6 kb germline deletion encompassing the promoter and first exon of NF1. This case report illustrates the relevance of WGS to detect structural variants including copy number variants that would be missed by alternative approaches. The identification of the causal pathogenic variant allowed a tailored genetic counseling with a targeted non-invasive prenatal diagnosis by detecting the deletion in plasmatic cell-free DNA from the proband’s pregnant partner. This report clearly highlights the need to make WGS a clinically accessible test, offering a tremendous opportunity to identify a molecular diagnosis for otherwise unsolved cases.


Similar content being viewed by others
Availability of data and materials
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
100,000 Genomes Project Pilot Investigators, Smedley D, Smith KR, et al (2021) 100,000 Genomes Pilot on Rare-Disease Diagnosis in Health Care—Preliminary Report. N Engl J Med 385:1868–1880. https://doi.org/10.1056/NEJMoa2035790
Bergqvist C, Servy A, Valeyrie-Allanore L et al (2020) Neurofibromatosis 1 French national guidelines based on an extensive literature review since 1966. Orphanet J Rare Dis 15:37. https://doi.org/10.1186/s13023-020-1310-3
Biesecker LG, Spinner NB (2013) A genomic view of mosaicism and human disease. Nat Rev Genet 14:307–320. https://doi.org/10.1038/nrg3424
Brems H, Beert E, de Ravel T, Legius E (2009) Mechanisms in the pathogenesis of malignant tumours in neurofibromatosis type 1. Lancet Oncol 10:508–515. https://doi.org/10.1016/S1470-2045(09)70033-6
Brems H, Chmara M, Sahbatou M et al (2007) Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet 39:1120–1126. https://doi.org/10.1038/ng2113
Brems H, Pasmant E, Van Minkelen R et al (2012) Review and update of SPRED1 mutations causing Legius syndrome. Hum Mutat 33:1538–1546. https://doi.org/10.1002/humu.22152
Castellanos E, Rosas I, Negro A et al (2020) Mutational spectrum by phenotype: panel-based NGS testing of patients with clinical suspicion of RASopathy and children with multiple café-au-lait macules. Clin Genet 97:264–275. https://doi.org/10.1111/cge.13649
Choi Y, Sims GE, Murphy S et al (2012) Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 7:e46688. https://doi.org/10.1371/journal.pone.0046688
Collins RL, Brand H, Karczewski KJ et al (2020) A structural variation reference for medical and population genetics. Nature 581:444–451. https://doi.org/10.1038/s41586-020-2287-8
Desmet F-O, Hamroun D, Lalande M et al (2009) Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. https://doi.org/10.1093/nar/gkp215
El Khattabi LA, Brun S, Gueguen P et al (2019) Performance of semiconductor sequencing platform for non-invasive prenatal genetic screening for fetal aneuploidy: results from a multicenter prospective cohort study in a clinical setting. Ultrasound Obstet Gynecol 54:246–254. https://doi.org/10.1002/uog.20112
Evans DG, Bowers N, Burkitt-Wright E et al (2016) Comprehensive RNA analysis of the NF1 gene in classically affected NF1 affected individuals meeting NIH criteria has high sensitivity and mutation negative testing is reassuring in isolated cases with pigmentary features only. EBioMedicine 7:212–220. https://doi.org/10.1016/j.ebiom.2016.04.005
Gruber A, Pacault M, El Khattabi LA et al (2018) Non-invasive prenatal diagnosis of paternally inherited disorders from maternal plasma: detection of NF1 and CFTR mutations using droplet digital PCR. Clin Chem Lab Med 56:728–738. https://doi.org/10.1515/cclm-2017-0689
Gutmann DH, Ferner RE, Listernick RH et al (2017) Neurofibromatosis type 1. Nat Rev Dis Primers 3:17004. https://doi.org/10.1038/nrdp.2017.4
Horan MP, Osborn M, Cooper DN, Upadhyaya M (2004) Functional analysis of polymorphic variation within the promoter and 5’ untranslated region of the neurofibromatosis type 1 (NF1) gene. Am J Med Genet A 131:227–231. https://doi.org/10.1002/ajmg.a.30358
Huby T, Le Guillou E, Burin des Roziers C et al (2021) Non-invasive prenatal diagnosis of a paternally inherited MEN1 pathogenic splicing variant. J Clin Endocrinol Metab. https://doi.org/10.1210/clinem/dgab894
Jian X, Boerwinkle E, Liu X (2014) In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 42:13534–13544. https://doi.org/10.1093/nar/gku1206
Kehrer-Sawatzki H, Cooper DN (2022) Challenges in the diagnosis of neurofibromatosis type 1 (NF1) in young children facilitated by means of revised diagnostic criteria including genetic testing for pathogenic NF1 gene variants. Hum Genet 141:177–191. https://doi.org/10.1007/s00439-021-02410-z
Kehrer-Sawatzki H, Mautner V-F, Cooper DN (2017) Emerging genotype–phenotype relationships in patients with large NF1 deletions. Hum Genet 136:349–376. https://doi.org/10.1007/s00439-017-1766-y
Legius E, Messiaen L, Wolkenstein P et al (2021) Revised diagnostic criteria for neurofibromatosis type 1 and Legius syndrome: an international consensus recommendation. Genet Med 23:1506–1513. https://doi.org/10.1038/s41436-021-01170-5
Louvrier C, Pasmant E, Briand-Suleau A et al (2018) Targeted next-generation sequencing for differential diagnosis of neurofibromatosis type 2, schwannomatosis, and meningiomatosis. Neuro Oncol 20:917–929. https://doi.org/10.1093/neuonc/noy009
Maertens O, De Schepper S, Vandesompele J et al (2007) Molecular dissection of isolated disease features in mosaic neurofibromatosis type 1. Am J Hum Genet 81:243–251. https://doi.org/10.1086/519562
Messiaen L, Yao S, Brems H et al (2009) Clinical and mutational spectrum of neurofibromatosis type 1-like syndrome. JAMA 302:2111–2118. https://doi.org/10.1001/jama.2009.1663
Messiaen LM, Callens T, Mortier G et al (2000) Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat 15:541–555. https://doi.org/10.1002/1098-1004(200006)15:6%3c541::AID-HUMU6%3e3.0.CO;2-N
Minoche AE, Lundie B, Peters GB et al (2021) ClinSV: clinical grade structural and copy number variant detection from whole genome sequencing data. Genome Med 13:32. https://doi.org/10.1186/s13073-021-00841-x
Pacot L, Vidaud D, Sabbagh A et al (2021) Severe phenotype in patients with large deletions of NF1. Cancers (Basel) 13:2963. https://doi.org/10.3390/cancers13122963
Pasmant E, de Saint-Trivier A, Laurendeau I et al (2008) Characterization of a 7.6-Mb germline deletion encompassing the NF1 locus and about a hundred genes in an NF1 contiguous gene syndrome patient. Eur J Hum Genet 16:1459–1466. https://doi.org/10.1038/ejhg.2008.134
Pasmant E, Parfait B, Luscan A et al (2015) Neurofibromatosis type 1 molecular diagnosis: what can NGS do for you when you have a large gene with loss of function mutations? Eur J Hum Genet 23:596–601. https://doi.org/10.1038/ejhg.2014.145
Pasmant E, Sabbagh A, Hanna N et al (2009a) SPRED1 germline mutations caused a neurofibromatosis type 1 overlap** phenotype. J Med Genet 46:425–430. https://doi.org/10.1136/jmg.2008.065243
Pasmant E, Sabbagh A, Masliah-Planchon J et al (2009b) Detection and characterization of NF1 microdeletions by custom high resolution array CGH. J Mol Diagn 11:524–529. https://doi.org/10.2353/jmoldx.2009.090064
Pasmant E, Vidaud M, Vidaud D, Wolkenstein P (2012) Neurofibromatosis type 1: from genotype to phenotype. J Med Genet 49:483–489. https://doi.org/10.1136/jmedgenet-2012-100978
Perez-Becerril C, Evans DG, Smith MJ (2021) Pathogenic noncoding variants in the neurofibromatosis and schwannomatosis predisposition genes. Hum Mutat 42:1187–1207. https://doi.org/10.1002/humu.24261
Leman R, Parfait B, Vidaud D et al (2022) SPiP, a comprehensive splicing prediction pipeline for massive detection of exonic and intronic variant effect on mRNA splicing. Hum Mutat. https://doi.org/10.22541/au.164544915.57104749/v1
Ratner N, Miller SJ (2015) A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15:290–301. https://doi.org/10.1038/nrc3911
Rentzsch P, Schubach M, Shendure J, Kircher M (2021) CADD-Splice-improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med 13:31. https://doi.org/10.1186/s13073-021-00835-9
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Robinson JT, Thorvaldsdóttir H, Winckler W et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Sabbagh A, Pasmant E, Imbard A et al (2013) NF1 molecular characterization and neurofibromatosis type I genotype-phenotype correlation: the French experience. Hum Mutat 34:1510–1518. https://doi.org/10.1002/humu.22392
Salomon LJ, Sotiriadis A, Wulff CB et al (2019) Risk of miscarriage following amniocentesis or chorionic villus sampling: systematic review of literature and updated meta-analysis. Ultrasound Obstet Gynecol 54:442–451. https://doi.org/10.1002/uog.20353
Trost B, Walker S, Wang Z et al (2018) A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am J Hum Genet 102:142–155. https://doi.org/10.1016/j.ajhg.2017.12.007
Yasunari T, Shiraki K, Hattori H, Miki T (2000) Frequency of choroidal abnormalities in neurofibromatosis type 1. Lancet 356:988–992. https://doi.org/10.1016/S0140-6736(00)02716-1
Yeo G, Burge CB (2004) Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11:377–394. https://doi.org/10.1089/1066527041410418
Acknowledgements
We thank all participants and their referring physicians.
Funding
No specific funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
All authors made substantial contributions to the work, as follows: V.P. and H.D. made substantial contributions to primary clinical data collection; M.V. supervised and performed the whole-genome analysis and variant validation; A.L., A.B-S., C.B.d.R. coordinated the whole-genome alignment, variant calling and variant interpretation; A.C., T.M., N.V., L.O., I.L., and C.B. made substantial contributions to molecular genetic experiments; N.H., D.H., B.P., L.E.K., J.N., and A.C. performed the genetic analyses; E.P., D.V., and L.P. substantially contributed to study concept and design and manuscript writing; analysis and interpretation of data as well as drafting of the manuscript; All authors provided critical review of the paper, have approved the submission, and accept responsibility for their contribution.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
The study is in line with the current French legislation on genetic studies.
Consent for publication
Patient and his parents gave their written informed consent for the use of their clinical and genetic data in the context of research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pacot, L., Pelletier, V., Chansavang, A. et al. Contribution of whole genome sequencing in the molecular diagnosis of mosaic partial deletion of the NF1 gene in neurofibromatosis type 1. Hum Genet 142, 1–9 (2023). https://doi.org/10.1007/s00439-022-02476-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-022-02476-3