Abstract
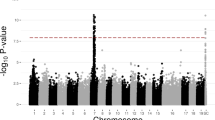
Dioecy has evolved numerous times in plants, but heteromorphic sex chromosomes are apparently rare. Sex determination has been studied in multiple Salix and Populus (Salicaceae) species, and P. trichocarpa has an XY sex determination system on chromosome 19, while S. suchowensis and S. viminalis have a ZW system on chromosome 15. Here we use whole genome sequencing coupled with quantitative trait locus map** and a genome-wide association study to characterize the genomic composition of the non-recombining portion of the sex determination region. We demonstrate that Salix purpurea also has a ZW system on chromosome 15. The sex determination region has reduced recombination, high structural polymorphism, an abundance of transposable elements, and contains genes that are involved in sex expression in other plants. We also show that chromosome 19 contains sex-associated markers in this S. purpurea assembly, along with other autosomes. This raises the intriguing possibility of a translocation of the sex determination region within the Salicaceae lineage, suggesting a common evolutionary origin of the Populus and Salix sex determination loci.






Similar content being viewed by others
References
Ainsworth C (2000) Boys and girls come out to play: the molecular biology of dioecious plants. Ann Bot 86:211–221. https://doi.org/10.1006/anbo.2000.1201
Alliende MC, Harper JL (1989) Demographic studies of a dioecious tree. I. Colonization, sex and age structure of a population of Salix cinerea. J Ecol 77:1029–1047. https://doi.org/10.2307/2260821
Alstrom-Rapaport C, Lascoux M, Wang YC et al (1998) Identification of a RAPD marker linked to sex determination in the basket willow (Salix viminalis L.). J Hered 89:44–49. https://doi.org/10.1093/jhered/89.1.44
Andrews KR, Good JM, Miller MR et al (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 17:81–92. https://doi.org/10.1038/nrg.2015.28
Arends D, Prins P, Jansen RC, Broman KW (2010) R/qtl: high-throughput multiple QTL map**. Bioinformatics 26:2990–2992
Argus GW (1997) Infrageneric classification of Salix (Salicaceae) in the new world. Syst Bot Monogr 52:1–121
Ashman T-L (2006) The evolution of separate sexes: a focus on the ecological context. In: Harder LD, Barrett SCH (eds) Ecology and evolution of flowers. Oxford University Press, Oxford, p 370
Ashman T-L, Kwok A, Husband BC (2013) Revisiting the Dioecy-Polyploidy Association: alternate pathways and research opportunities. Cytogenet Genome Res 140:241–255. https://doi.org/10.1159/000353306
Bachtrog D, Mank J, Peichel CL et al (2014) Sex determination: why so many ways of doing it? PLoS Biol 12:e1001899. https://doi.org/10.1371/journal.pbio.1001899
Barrett SCH, Hough J (2013) Sexual dimorphism in flowering plants. J Exp Bot 64:67–82. https://doi.org/10.1093/jxb/err313
Bergero R, Charlesworth D (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102
Bergero R, Forrest A, Kamau E, Charlesworth D (2007) Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175:1945–1954. https://doi.org/10.1534/genetics.106.070110
Beukeboom LW, Perrin N (2014) The evolution of sex determination. Oxford University Press, New York
Boucher LD, Manchester SR, Judd WS (2003) An extinct genus of Salicaceae based on twigs with attached flowers, fruits, and foliage from the Eocene Green River Formation of Utah and Colorado, USA. Am J Bot 90:1389–1399. https://doi.org/10.3732/ajb.90.9.1389
Bull JJ, Charnov EL (1977) Changes in the heterogametic mechanism of sex determination. Heredity 39:1–14. https://doi.org/10.1038/hdy.1977.38
Burby PE, Simmons LA (2017) MutS2 promotes homologous recombination in Bacillus subtilis. J Bacteriol 199:e00682–e00616. https://doi.org/10.1128/JB.00682-16
Cai G, Cresti M (2009) Organelle motility in the pollen tube: a tale of 20 years. J Exp Bot 60:495–508. https://doi.org/10.1093/jxb/ern321
Carlson CH, Choi Y, Chan AP et al (2017) Dominance and Sexual Dimorphism Pervade the Salix purpurea L. Transcriptome. Genome Biol Evol 9:2377–2394. https://doi.org/10.1093/gbe/evx174
Charlesworth D (2006) Evolution of plant breeding systems. Curr Biol 16:726–735. https://doi.org/10.1016/j.cub.2006.07.068
Charlesworth D (2015) Plant contributions to our understanding of sex chromosome evolution. New Phytol 208:52–65. https://doi.org/10.1111/nph.13497
Charlesworth D (2016) Plant sex chromosomes. Annu Rev Plant Biol 67:397–420. https://doi.org/10.1146/annurev-arplant-043015-111911
Charlesworth D, Charlesworth B (1978) Population genetics of partial male-sterility and the evolution of monoecy and dioecy. Heredity 41:137–153. https://doi.org/10.1038/hdy.1978.83
Charnov EL (1982) The theory of sex allocation. Monogr Popul Biol 18:1–355. https://doi.org/10.1017/CBO9781107415324.004
Chen Y, Wang T, Fang L et al (2016) Confirmation of single-locus sex determination and female heterogamety in willow based on linkage analysis. PLoS One 11:e0147671. https://doi.org/10.1371/journal.pone.0147671
DePristo M, Banks E, Poplin R et al (2011) A framework for variation discovery and genoty** using next-generation DNA sequencing data. Nat Genet 43:491–498. https://doi.org/10.1038/ng.806
Dickmann DI, Kuzovkina J (2014) Poplars and willows of the world, with emphasis on silviculturally important species. In: Poplars and willows: trees for society and the environment. CABI, Wallingford, pp 8–91
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genoty**-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Falda M, Toppo S, Pescarolo A et al (2012) Argot2: a large scale function prediction tool relying on semantic similarity of weighted Gene Ontology terms. BMC Bioinformatics 13:S14. https://doi.org/10.1186/1471-2105-13-S4-S14
Fukui K, Kuramitsu S (2011) Structure and function of the small MutS-related domain. Mol Biol Int 2011:1–9. https://doi.org/10.4061/2011/691735
Fukui K, Kosaka H, Kuramitsu S, Masui R (2007) Nuclease activity of the MutS homologue MutS2 from Thermus thermophilus is confined to the Smr domain. Nucleic Acids Res 35:850–860. https://doi.org/10.1093/nar/gkl735
Füssel U, Dötterl S, Jürgens A, Aas G (2007) Inter- and intraspecific variation in floral scent in the genus Salix and its implication for pollination. J Chem Ecol 33:749–765. https://doi.org/10.1007/s10886-007-9257-6
Gaudet M, Jorge V, Paolucci I et al (2008) Genetic linkage maps of Populus nigra L. including AFLPs, SSRs, SNPs, and sex trait. Tree Genet Genomes 4:25–36. https://doi.org/10.1007/s11295-007-0085-1
Geraldes A, Hefer CA, Capron A et al (2015) Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol 24:3243–3256. https://doi.org/10.1111/mec.13126
Glaubitz JC, Casstevens TM, Lu F et al (2014) TASSEL-GBS: a high capacity genoty** by sequencing analysis pipeline. PLoS One 9:e0090346. https://doi.org/10.1371/journal.pone.0090346
Glick L, Sabath N, Ashman TL et al (2016) Polyploidy and sexual system in angiosperms: is there an association? Am J Bot 103:1223–1235. https://doi.org/10.3732/ajb.1500424
Gnerre S, MacCallum I, Przybylski D et al (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci USA 108:1513–1518. https://doi.org/10.1073/pnas.1017351108
Goodstein DM, Shu S, Howson R et al (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186. https://doi.org/10.1093/nar/gkr944
Hou J, Ye N, Zhang D et al (2015) Different autosomes evolved into sex chromosomes in the sister genera of Salix and Populus. Sci Rep 5:e9076. https://doi.org/10.1038/srep09076
Hou J, Ye N, Dong Z et al (2016) Major chromosomal rearrangements distinguish willow and poplar after the ancestral “Salicoid” genome duplication. Genome Biol Evol 8:1868–1875. https://doi.org/10.1093/gbe/evw127
Kang HM, Sul JH, Service SK et al (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42:348–354. https://doi.org/10.1038/ng.548
Karp A, Hanley SJ, Trybush SO et al (2011) Genetic Improvement of Willow for Bioenergy and Biofuels. J Integr Plant Biol 53:151–165. https://doi.org/10.1111/j.1744-7909.2010.01015.x
Karrenberg S, Kollmann J, Edwards PJ (2002) Pollen vectors and inflorescence morphology in four species of Salix. Plant Syst Evol 235:181–188. https://doi.org/10.1007/s00606-002-0231-z
Kersten B, Pakull B, Groppe K et al (2014) The sex-linked region in Populus tremuloides Turesson 141 corresponds to a pericentromeric region of about two million base pairs on P. trichocarpa chromosome 19. Plant Biol 16:411–418. https://doi.org/10.1111/plb.12048
Kunkel T, Erie D (2005) DNA mismatch repair. Annu Rev Biochem 74:681–710. https://doi.org/10.1146/annurev.biochem.74.082803.133243
Lian S, Liu T, Gong K et al (2016) A complete and accurate short sequence alignment algorithm for repeats. J Biosci Med 04:144–151. https://doi.org/10.4236/jbm.2016.412018
Lin H, Niu L, McHale N et al (2013) Evolutionarily conserved repressive activity of WOX proteins mediates leaf blade outgrowth and floral organ development in plants. Proc Natl Acad Sci USA 110:366–371. https://doi.org/10.1073/pnas.1215376110
Lloyd DG (1979) Evolution towards dioecy in heterostylous populations. Plant Syst Evol 131:71–80. https://doi.org/10.1007/BF00984123
Mank JE (2009) Sex chromosomes and the evolution of sexual dimorphism: lessons from the genome. Am Nat 173:141–150. https://doi.org/10.1086/595754
Melters DP, Bradnam KR, Young H et al (2013) Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol 14:R10. https://doi.org/10.1186/gb-2013-14-1-r10
Miller JR, Koren S, Sutton G (2010) Assembly algorithms for next-generation sequencing data. Genomics 95:315–327. https://doi.org/10.1016/j.ygeno.2010.03.001
Ming R, Moore PH (2007) Genomics of sex chromosomes. Curr Opin Plant Biol 10:123–130. https://doi.org/10.1016/j.pbi.2007.01.013
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514. https://doi.org/10.1146/annurev-arplant-042110-103914
Mock KE, Callahan CM, Islam-Faridi MN et al (2012) Widespread triploidy in western North American aspen (Populus tremuloides). PLoS One 7:e48406. https://doi.org/10.1371/journal.pone.0048406
Moore EC, Roberts RB (2013) Polygenic sex determination. Curr Biol 23:R510–R512. https://doi.org/10.1016/j.cub.2013.04.004
Nicolas M, Marais G, Hykelova V et al (2005) A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol 3:e4. https://doi.org/10.1371/journal.pbio.0030004
Nishihama R, Soyano T, Ishikawa M et al (2002) Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109:87–99. https://doi.org/10.1016/S0092-8674(02)00691-8
Olson MS, Hamrick JL, Moore RC (2017) Breeding systems, mating systems, and gender determination in angiosperm trees. In: Groover A, Cronk QCB (eds) Comparative and evolutionary genomics of angiosperm trees. Springer International Publishing, Switzerland, pp 139–158
Otto SP, Pannell JR, Peichel CL et al (2011) About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet 27:358–367. https://doi.org/10.1016/j.tig.2011.05.001
Pakull B, Groppe K, Meyer M et al (2009) Genetic linkage map** in aspen (Populus tremula L. and Populus tremuloides Michx.). Tree Genet Genomes 5:505–515. https://doi.org/10.1007/s11295-009-0204-2
Pakull B, Kersten B, Lüneburg J, Fladung M (2014) A simple PCR-based marker to determine sex in aspen. Plant Biol 17:256–261. https://doi.org/10.1111/plb.12217
Pandey RS, Azad RK (2016) Deciphering evolutionary strata on plant sex chromosomes and fungal mating-type chromosomes through compositional segmentation. Plant Mol Biol 90:359–373. https://doi.org/10.1007/s11103-015-0422-y
Paolucci I, Gaudet M, Jorge V et al (2010) Genetic linkage maps of Populus alba L. and comparative map** analysis of sex determination across Populus species. Tree Genet Genomes 6:863–875. https://doi.org/10.1007/s11295-010-0297-7
Peto FH (1938) Cytology of poplar species and natural hybrids. Can J Res 16:446–455
Price AL, Patterson NJ, Plenge RM et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. https://doi.org/10.1038/ng1847
Pucholt P, Rönnberg-Wästljung A-C, Berlin S (2015) Single locus sex determination and female heterogamety in the basket willow (Salix viminalis L.). Heredity 114:575–583. https://doi.org/10.1038/hdy.2014.125
Pucholt P, Hallingbäck HR, Berlin S (2017a) Allelic incompatibility can explain female biased sex ratios in dioecious plants. BMC Genom 18:251. https://doi.org/10.1186/s12864-017-3634-5
Pucholt P, Wright AE, Conze LL et al (2017b) Recent sex chromosome divergence despite ancient Dioecy in the Willow, Salix viminalis. Mol Biol Evol 22:522–525. https://doi.org/10.1093/molbev/msx144
Qi J, Chen Y, Copenhaver GP, Ma H (2014) Detection of genomic variations and DNA polymorphisms and impact on analysis of meiotic recombination and genetic map**. Proc Natl Acad Sci 111:10007–10012. https://doi.org/10.1073/pnas.1321897111
Renner SS (2014) The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am J Bot 101:1588–1596. https://doi.org/10.3732/ajb.1400196
Rice WWR (1984) Sex chromosomes and the evolution of sexual dimorphism. Evolution 38:1416–1424. https://doi.org/10.2307/2408385
Serapiglia MJ, Gouker FE, Hart JF et al (2015) Ploidy level affects important biomass traits of novel shrub Willow (Salix) hybrids. BioEnergy Res 8:259–269. https://doi.org/10.1007/s12155-014-9521-x
Slavov GT, Zhelev P (2010) Salient biological features, systematics, and genetic variation of Populus. In: Jansson S, Bhalerao RP, Groover A (eds) Genetics and genomics of Populus. Springer, New York, pp 15–38
Sterck L, Rombauts S, Jansson S et al (2005) EST data suggest that poplar is an ancient polyploid. New Phytol 167:165–170. https://doi.org/10.1111/j.1469-8137.2005.01378.x
Temmel NA, Rai HS, Cronk QCB (2007) Sequence characterization of the putatively sex-linked Ssu72 -like locus in willow and its homologue in poplar. Can J Bot 85:1092–1097. https://doi.org/10.1139/B07-058
Tuskan GA, DiFazio S, Jansson S et al (2006) The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313:1596–1604. https://doi.org/10.1126/science.1128691
Tuskan GA, DiFazio S, Faivre-Rampant P et al (2012) The obscure events contributing to the evolution of an incipient sex chromosome in Populus: a retrospective working hypothesis. Tree Genet Genomes 8:559–571. https://doi.org/10.1007/s11295-012-0495-6
Ueno N, Suyama Y, Seiwa K (2007) What makes the sex ratio female-biased in the dioecious tree Salix sachalinensis? J Ecol 95:951–959. https://doi.org/10.1111/j.1365-2745.2007.01269.x
van Doorn GS, Kirkpatrick M (2007) Turnover of sex chromosomes induced by sexual conflict. Nature 449:909–912. https://doi.org/10.1038/nature06178
van Doorn GS, Kirkpatrick M (2010) Transitions between male and female heterogamety caused by sex-antagonistic selection. Genetics 186:629–645
Vyskot B, Hobza R (2015) The genomics of plant sex chromosomes. Plant Sci 236:126–135. https://doi.org/10.1016/j.plantsci.2015.03.019
Wang J, Na J, Yu Q et al (2012) Sequencing papaya X and Y h chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci 109:13710–13715. https://doi.org/10.1073/pnas.1207833109/-/DCSupplemental.www.pnas.org/cgi/doi/10.1073/pnas.1207833109
Westergaard M (1958) The mechanism of sex determination in dioecious flowering plants. Adv Genet 9:217–281. https://doi.org/10.1016/S0065-2660(08)60163-7
Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. https://doi.org/10.1093/molbev/msm088
Yin T, DiFazio SP, Gunter LE et al (2008) Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res 18:422–430. https://doi.org/10.1101/gr.7076308
Zhou W, Lu Q, Li Q et al (2017) PPR-SMR protein SOT1 has RNA endonuclease activity. Proc Natl Acad Sci USA 114:E1554–E1563. https://doi.org/10.1073/pnas.1612460114
Acknowledgements
We are grateful to Matt Olson for helpful comments on the manuscript.
Funding
This work was supported by grants from the USDA-NIFA CAP program (4705-WVU-USDA-9703), the DOE JGI Community Sequencing Program, and the NSF Dimensions of Biodiversity Program (DEB-1542509). Sequencing was conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported by the Office of Science of the U.S. Department of Energy under Contract no. DE-AC02-05CH11231.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Data availability
All raw sequencing data are available from the NCBI Sequence Read Archive (accessions SRP003908, SRP086434, and SRP086435) and the genome assembly and annotation are available from Phytozome (https://phytozome.jgi.doe.gov).
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2018_1473_MOESM1_ESM.xlsx
Table S1 Significant markers (LOD>3.5) from QTL map** of sex. The table includes linkage group (LG), map positions (in centimorgans), map type (female backcross, F, male backcross, M, and intercross, IC), the physical scaffold from the genome assembly, the physical position of the marker in the genome assembly, and the frequency of different genotype configurations in the progeny. Table S2 Number of unfiltered GBS markers produced by the Tassel pipeline for the F2 family 317. Markers/100kb is the average number of markers per 100 kb interval. F:M Backcross is the ratio of markers in a Female Backcross configuration (heterozygous in the female parent, homozygous in the male parent) to markers in the Male Backcross configuration (homozygous in female parent, heterozygous in male parent). Table S3 Results of GWAS for sex. The table includes all significant markers (p<1x10-7). Table S4 Best matches for secondary S. purpurea SDRs to the S. purpurea and P. trichocarpa genomes. “Secondary Blast Hit” is the best blastn hit to the S. purpurea genome, after excluding self hits. Table S5 Markers showing a female-specific genotype configuration (one allele observed in females, none in males). These are presumably derived from W segments included in the genome assembly. Table S6 Scaffolds with >30% female-specific sequence. “Proportion W” is a calculation based on the proportion of the scaffold, after excluding gaps, that is present in the female sequence but absent in the male sequence (Female-Specific). Table S7 Repeat composition of the S. purpurea chromosomes. Table S8 Predicted genes found within the SDR of S. purpurea. “W Overlap” and “W proportion” represent the intersection of the location of the gene with female-specific genome segments. Omega values are the ratio of nonsynonymous (dN) to synonymous (dS) substitutions between the S. purpurea and P. trichocarpa orthologs. Multiple values are provided in cases with multiple Populus orthologs, presumably due to lineage-specific expansion (XLSX 148 KB)
438_2018_1473_MOESM2_ESM.pdf
Fig. S1 Pairwise Scaled Identity by State (IBS) for the (a) complete association population (N=112), (b) the complete F2 full sib Family (N=497), and (c) the association population with clones removed (N=75). The IBS cutoff used for identifying clonal pairs was 0.9. Fig. S2 Frequency of mapped markers with and without segregation distortion in family 317 for males and females. A. Markers in female-backcross configuration. B. Markers in male-backcross configuration. Notice the lack of undistorted (normal) markers on chr 19 in female backcross configuration. Fig. S3 Quantile–Quantile (Q–Q) plots of observed and expected P-values for the GWAS for sex. Red line indicates X = Y. Fig. S4 Box plots of average observed heterozygosity for males and females for sex-associated loci in the S. purpurea association population. Fig. S5 Distribution of differences in null allele frequency between females and males in the association population. Extreme values are shaded in red. Fig. S6 Proportion of reference sequence gaps (“assembly Ns”) in regions that showed no coverage in the female (a) or male (b) reference-based alignments. The male had 0 coverage primarily in regions with minimal reference gaps, suggesting that these are regions that are present in the female sequence and absent in the male. Fig. S7 Box plot showing that the proportion of repeat elements is elevated in the SDR. Fig. S8 Delineation of putative centromeres relative to the SDRs, for chromosomes not shown in the main text. Bar plots represent, from the top, gene density, repeat density, density of centromeric repeats, and physical:genetic distance ratio (Mb/cM) in 100 kb windows. Blue shading shows positions of putative centromeres, as defined by empirical thresholds represented by horizontal red lines. The position of the SDRs are indicated by vertical red shading. Fig. S9 Box plots comparing the composition of putative centromeric intervals to the rest of the genome, including (from top to bottom) gene content, total repeat content, presence of putative centromere-associated repeat elements, and physical:genetic distance ratio (Mb/cM). Fig. S10 Dot plot derived from aligning the S. suchowensis SDR (primarily located on scaffold64) to S. purpurea chr 15 using lastz. Fig. S11 Alignment of Kinesin genes from the SDR of S. purpurea and their closest ortholog in P. trichocarpa. SapurV1A.1267s0010 is artificially truncated due to an assembly gap overlap** with the gene. Conserved domains are highlighted and labeled. Tandem duplicate pairs are 1.) SapurV1A.0719s0080 and SapurV1A.0719s0090; and 2.) SapurV1A.1267s0010 and SapurV1A.1267s0020 (PDF 4560 KB)
Rights and permissions
About this article
Cite this article
Zhou, R., Macaya-Sanz, D., Rodgers-Melnick, E. et al. Characterization of a large sex determination region in Salix purpurea L. (Salicaceae). Mol Genet Genomics 293, 1437–1452 (2018). https://doi.org/10.1007/s00438-018-1473-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1473-y