Abstract
Main conclusion
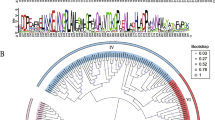
Sixteen Lhc genes representing 13 phylogenetic groups were identified from the full-length transcriptome of tigernut, exhibiting development regulation and diurnal fluctuation expression patterns in leaves.
Abstract
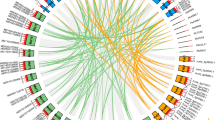
Nuclear encoded light-harvesting chlorophyll a/b-binding (Lhc) proteins play indispensable roles in oxygenic photosynthesis. In this study, we present the first transcriptome-based characterization of Lhc family genes in tigernut (Cyperus esculentus L.), a Cyperaceae C4 plant producing oil in underground tubers. A number of 16 Lhc genes representing 13 phylogenetic groups identified from the full-length tigernut transcriptome are equal to that found in both Carex littledalei (another Cyperaceae plant) and papaya, slightly more than 15 members present in both rice and jatropha, but relatively less than 18, 20, and 21 members present in sorghum, cassava, and Arabidopsis, respectively. Nevertheless, nearly one-vs-one orthologous relationship was observed in most groups, though some of them are no longer located in syntenic blocks and species-specific expansion was frequently found in Lhcb1. Comparative genomics analysis revealed that the loss of two groups (i.e., Lhca2 and Lhca5) in C. littledalei is species-specific, sometime after the split with tigernut, and the expansion of Lhcb1 was mainly contributed by tandem duplication as observed in most species. Interestingly, a transposed duplication, which appears to be shared by monocots, was also identified in Lhcb1. Further transcriptome profiling revealed a predominant expression pattern of most CeLhc family genes in photosynthetic tissues and enhanced transcription during leaf maturation, reflecting their key roles in light absorption. Moreover, qRT-PCR analysis revealed an apparent diurnal fluctuation expression pattern of 11 dominant CeLhc genes. These findings not only highlight species-specific evolution of Lhc genes in the Cyperaceae family as well as the monocot lineage, but also provide valuable information for further functional analysis and genetic improvement in tigernut.






Similar content being viewed by others
Data availability
Data availability All data analysed during this study are included in Suppl. Table S2.
Abbreviations
- CB:
-
Chlorophyll a/b-binding
- Lhc:
-
Light-harvesting chlorophyll a/b-binding
- Ka:
-
Non-synonymous substitution rate
- Ks:
-
Synonymous substitution rate
- OG:
-
Orthogroup
- WGD:
-
Whole-genome duplication
References
Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T (2008) In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE 3:e2033. https://doi.org/10.1371/journal.pone.0002033
Altschul SF, Madden TL, Schaffer AA et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. https://doi.org/10.1093/nar/25.17.3389
Andersson J, Walters RG, Horton P, Jansson S (2001) Antisense inhibition of the photosynthetic antenna proteins CP29 and CP26: implications for the mechanism of protective energy dissipation. Plant Cell 13:1193–1204. https://doi.org/10.1105/tpc.13.5.1193
Andersson J, Wentworth M, Walters RG et al (2003) Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II - effects on photosynthesis, grana stacking and fitness. Plant J 35:350–361. https://doi.org/10.1046/j.1365-313x.2003.01811.x
Bai X, Chen T, Wu Y et al (2021) Selection and validation of reference genes for qRT-PCR analysis in the oil-rich tuber crop tiger nut (Cyperus esculentus) based on transcriptome data. Int J Mol Sci. https://doi.org/10.3390/ijms22052569
Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426:630–635. https://doi.org/10.1038/nature02200
de Bianchi S, Dall’Osto L, Tognon G, Morosinotto T, Bassi R (2008) Minor antenna proteins CP24 and CP26 affect the interactions between photosystem II subunits and the electron transport rate in grana membranes of Arabidopsis. Plant Cell 20:1012–1028. https://doi.org/10.1105/tpc.107.055749
de Bianchi S, Betterle N, Kouril R et al (2011) Arabidopsis mutants deleted in the light-harvesting protein Lhcb4 have a disrupted photosystem II macrostructure and are defective in photoprotection. Plant Cell 23:2659–2679. https://doi.org/10.1105/tpc.111.087320
Bowers JE, Chapman BA, Rong J, Paterson AH (2003) Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422:433–438. https://doi.org/10.1038/nature01521
Broglie R, Bellemare G, Bartlett SG, Chua NH, Cashmore AR (1981) Cloned DNA sequences complementary to mRNAs encoding precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase and a chlorophyll a/b binding polypeptide. Proc Natl Acad Sci USA 78:7304–7308. https://doi.org/10.1073/pnas.78.12.7304
Brunner H, Ruediger W (1995) The greening process in cress seedlings IV. Light regulated expression of single Lhc genes. J Photochem Photobiol B 27:257–263
Can M, Wei W, Zi H et al (2020) Genome sequence of Kobresia littledalei, the first chromosome-level genome in the family Cyperaceae. Sci Data 7:175. https://doi.org/10.1038/s41597-020-0518-3
Chen C, Chen H, Zhang Y et al (2020) TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Codina-Torrella I, Guamis B, Trujillo AJ (2015) Characterization and comparison of tiger nuts (Cyperus esculentus L.) from different geographical origin. Ind Crop Prod 65:406–414. https://doi.org/10.1016/j.indcrop.2014.11.007
Coruzzi G, Broglie R, Cashmore A, Chua NH (1983) Nucleotide sequences of two pea cDNA clones encoding the small subunit of ribulose 1,5-bisphosphate carboxylase and the major chlorophyll a/b-binding thylakoid polypeptide. J Biol Chem 258:1399–1402
Croce R, van Amerongen H (2020) Light harvesting in oxygenic photosynthesis: Structural biology meets spectroscopy. Science 369:eaay2058. https://doi.org/10.1126/science.aay2058
De Castro O, Gargiulo R, Del Guacchio E, Caputo P, De Luca P (2015) A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, Poales): two sides of the same coin (weed vs. crop). Ann Bot 115:733–745. https://doi.org/10.1093/aob/mcv001
Dall'Osto L, Ünlü C, Cazzaniga S, van Amerongen H (2014) Disturbed excitation energy transfer in Arabidopsis thaliana mutants lacking minor antenna complexes of photosystem II. Biochim Biophys Acta. 1837:1981–1988. https://doi.org/10.1016/j.bbabio.2014.09.011
Defelice MS (2002) Yellow nutsedge Cyperus esculentus L.—Snack food of the gods. Weed Technol 16:901–907. https://doi.org/10.1614/0890-037X(2002)016[0901:YNCELS]2.0.CO;2
Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Elrad D, Grossman AR (2004) A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr Genet 45:61–75. https://doi.org/10.1007/s00294-003-0460-x
Engelken J, Brinkmann H, Adamska I (2010) Taxonomic distribution and origins of the extended LHC (light-harvesting complex) antenna protein superfamily. BMC Evol Biol 10:233. https://doi.org/10.1186/1471-2148-10-233
Grossman AR, Bhaya D, Apt KE, Kehoe DM (1995) Light-harvesting complexes in oxygenic photosynthesis: diversity, control, and evolution. Annu Rev Genet 29:231–288. https://doi.org/10.1146/annurev.ge.29.120195.001311
Hobe S, Förster R, Klingler J, Paulsen H (1995) N-proximal sequence motif in light-harvesting chlorophyll a/b-binding protein is essential for the trimerization of light-harvesting chlorophyll a/b complex. Biochemistry 34:10224–10228. https://doi.org/10.1021/bi00032a016
Ilíková I, Ilík P, Opatíková M et al (2021) Towards spruce-type photosystem II: consequences of the loss of light-harvesting proteins LHCB3 and LHCB6 in Arabidopsis. Plant Physiol 187:2691–2715. https://doi.org/10.1093/plphys/kiab396
Jansson S (1999) A guide to the Lhc genes and their relatives in Arabidopsis. Trends Plant Sci 4:236–240. https://doi.org/10.1016/s1360-1385(99)01419-3
Jiao Y, Leebens-Mack J, Ayyampalayam S et al (2012) A genome triplication associated with early diversification of the core eudicots. Genome Biol 13:R3. https://doi.org/10.1186/gb-2012-13-1-r3
Jiao Y, Li J, Tang H, Paterson AH (2014) Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26:2792–2802. https://doi.org/10.1105/tpc.114.127597
Klimmek F, Sjödin A, Noutsos C, Leister D, Jansson S (2006) Abundantly and rarely expressed Lhc protein genes exhibit distinct regulation patterns in plants. Plant Physiol 140:793–804. https://doi.org/10.1104/pp.105.073304
Kloppstech K (1985) Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta 165:502–506. https://doi.org/10.1007/BF00398095
Koziol AG, Borza T, Ishida K et al (2007) Tracing the evolution of the light-harvesting antennae in chlorophyll a/b-containing organisms. Plant Physiol 143:1802–1816. https://doi.org/10.1104/pp.106.092536
Kühlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367:614–621. https://doi.org/10.1038/367614a0
Larridon I, Reynders M, Huygh W et al (2011) Affinities in C3 Cyperus lineages (Cyperaceae) revealed using molecular phylogenetic data and carbon isotope analysis. Bot J Lin Soc 167:19–46. https://doi.org/10.1111/j.1095-8339.2011.01160.x
Li XW, Zhu YL, Chen CY et al (2020) Cloning and characterization of two chlorophyll A/B binding protein genes and analysis of their gene family in Camellia sinensis. Sci Rep 10:4602. https://doi.org/10.1038/s41598-020-61317-3
Liu Z, Yan H, Wang K et al (2004) Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 428:287–292. https://doi.org/10.1038/nature02373
Luo J, Abid M, Tu J et al (2022) Genome-wide identification of the LHC gene family in kiwifruit and regulatory role of AcLhcb3.1/3.2 for chlorophyll a content. Int J Mol Sci 23:6528. https://doi.org/10.3390/ijms23126528
Maduka N, Ire FS (2018) Tigernut plant and useful application of tigernut tubers (Cyperus esculentus) - a review. Cur J App Sci Technol 29(3):1–23. https://doi.org/10.9734/CJAST/2018/43551
Makareviciene V, Gumbytea M, Yunik A et al (2013) Opportunities for the use of chufa sedge in biodiesel production. Ind Crop Prod 50:633–637. https://doi.org/10.1016/j.indcrop.2013.08.036
Nelson N, Junge W (2015) Structure and energy transfer in photosystems of oxygenic photosynthesis. Annu Rev Biochem 84:659–683. https://doi.org/10.1146/annurev-biochem-092914-041942
Niu L, Zhang Y, Yang C et al (2022) Complete mitochondrial genome sequence and comparative analysis of the cultivated yellow nutsedge. Plant Genome 22:e20239. https://doi.org/10.1002/tpg2.20239
Pan X, Li M, Wan T et al (2011) Structural insights into energy regulation of light-harvesting complex CP29 from spinach. Nat Struct Mol Biol 18:309–315. https://doi.org/10.1038/nsmb.2008
Pichulla B (1988) Plastid and nuclear mRNA fluctuations in tomato leaves - diurnal and circadian rhythms during extended dark and light periods. Plant Mol Biol 11:345–353. https://doi.org/10.1126/10.1007/BF00027391
Pietrzykowska M, Suorsa M, Semchonok DA et al (2014) The light-harvesting chlorophyll a/b binding proteins Lhcb1 and Lhcb2 play complementary roles during state transitions in Arabidopsis. Plant Cell 26:3646–3660. https://doi.org/10.1105/tpc.114.127373
Qiao X, Li Q, Yin H et al (2019) Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol 20:38. https://doi.org/10.1186/s13059-019-1650-2
Qin X, Suga M, Kuang T, Shen JR (2015) Photosynthesis. atructural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 348:989–995. https://doi.org/10.1126/science.aab0214
Ren W, Guo D, **ng G et al (2021) Complete chloroplast genome sequence and comparative and phylogenetic analyses of the cultivated Cyperus esculentus. Diversity 13:405. https://doi.org/10.3390/d13090405
Roalson EH (2008) A synopsis of chromosome number variation in the Cyperaceae. Bot Rev 74:209–393. https://doi.org/10.1007/s12229-008-9011-y
Rogl H, Kühlbrandt W (1999) Mutant trimers of light-harvesting complex II exhibit altered pigment content and spectroscopic features. Biochemistry 38:16214–16222. https://doi.org/10.1021/bi990739p
Sattari Vayghan H, Nawrocki WJ et al (2022) Photosynthetic light harvesting and thylakoid organization in a CRISPR/Cas9 Arabidopsis thaliana LHCB1 knockout mutant. Front Plant Sci 13:833032. https://doi.org/10.3389/fpls.2022.833032
Standfuss J, Terwisscha van Scheltinga AC, Lamborghini M, Kühlbrandt W (2005) Mechanisms of photoprotection and nonphotochemical quenching in pea light-harvesting complex at 2.5 a resolution. EMBO J 24:919–928. https://doi.org/10.1038/sj.emboj.7600585
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tobin EM (1981) Photochrome-mediated regulation of messenger RNAs for the small subunit of ribulose 1, 5-bisphosphate carboxylase and the light-harvesting chlorophyll a/b-protein in Lemna gibba. Plant Mol Biol 1:35–51. https://doi.org/10.1007/BF00023012
Turesson H, Marttila S, Gustavsson KE et al (2010) Characterization of oil and starch accumulation in tubers of Cyperus esculentus var. sativus (Cyperaceae): a novel model system to study oil reserves in nonseed tissues. Am J Bot 97:1884–1893. https://doi.org/10.3732/ajb.1000200
Wang B, Tseng E, Regulski M et al (2016) Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat Commun 7:11708. https://doi.org/10.1038/ncomms11708
Wientjes E, Oostergetel GT, Jansson S, Boekema EJ, Croce R (2009) The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. J Biol Chem 284:7803–7810. https://doi.org/10.1074/jbc.M808395200
**ao YH, Zou Z, Zhao YG, Guo AP, Zhang L (2022) Molecular cloning and characterization of an acetolactate synthase gene (CeALS) from tigernut (Cyperus esculentus L.). Biotech Bull 38:184–192. https://doi.org/10.3560/j.cnki.biotech.bull.1985.2021-1198
Xu P, Roy LM, Croce R (2017) Functional organization of photosystem II antenna complexes: CP29 under the spotlight. Biochim Biophys Acta Bioenerg 1858:815–822. https://doi.org/10.1016/j.bbabio.2017.07.003
Xu S, Zou Z, **ao YH et al (2022) Cloning and functional characterization of CeWRI1, a gene involved in oil accumulation from tigernut (Cyperus esculentus L.) tubers. Chin J Trop Crop 43:923–929
Yang X, Niu L, Zhang Y et al (2022) Morpho-agronomic and biochemical characterization of accessions of tiger nut (Cyperus esculentus) grown in the north temperate zone of China. Plants (basel) 11:923. https://doi.org/10.3390/plants11070923
Yu Y, Lu X, Zhang T et al (2022) Tiger nut (Cyperus esculentus L): Nutrition, processing, function and applications. Foods 11:601
Zhang Q, Ma C, Wang X et al (2021) Genome-wide identification of the light-harvesting chlorophyll a/b-binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb2.3 on chlorophyll a synthesis. Plant Biol (stuttg) 23:831–842. https://doi.org/10.1111/plb.13294
Zhang S, Li P, Wei Z et al (2022) Cyperus (Cyperus esculentus L.): a review of its compositions, medical efficacy, antibacterial activity and allelopathic potentials. Plants (basel) 11:1127. https://doi.org/10.3390/plants11091127
Zhao S, Gao H, Luo J et al (2020a) Genome-wide analysis of the light-harvesting chlorophyll a/b-binding gene family in apple (Malus domestica) and functional characterization of MdLhcb4.3, which confers tolerance to drought and osmotic stress. Plant Physiol Biochem 154:517–529. https://doi.org/10.1016/j.plaphy.2020.06.022
Zhao YG, Kong H, Guo YL, Zou Z (2020b) Light-harvesting chlorophyll a/b-binding protein-encoding genes in jatropha and the comparison with castor, cassava and arabidopsis. PeerJ 8:e8465. https://doi.org/10.7717/peerj.8465
Zou Z, Yang JH (2019) Genomics analysis of the light-harvesting chlorophyll a/b-binding (Lhc) superfamily in cassava (Manihot esculenta Crantz). Gene 702:171–181. https://doi.org/10.1016/j.gene.2019.03.071
Zou Z, Gong J, An F et al (2015) Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genom 16:1001. https://doi.org/10.1186/s12864-015-2152-6
Zou Z, **e G, Yang L (2017) Papain-like cysteine protease encoding genes in rubber (Hevea brasiliensis): Comparative genomics, phylogenetic, and transcriptional profiling analysis. Planta 246:999–1018. https://doi.org/10.1007/s00425-017-2739-z
Zou Z, Li MY, Jia RZ et al (2020) Genes encoding light-harvesting chlorophyll a/b-binding proteins in papaya (Carica papaya L.) and insight into lineage-specific evolution in Brassicaceae. Gene 748:144685. https://doi.org/10.1016/j.gene.2020.144685
Zou Z, Zhao YG, Zhang L, Kong H, Guo YL, Guo AP (2021) Single-molecule real-time (SMRT)-based full-length transcriptome analysis of tigernut (Cyperus esculentus L.). Chin J Oil Crop Sci. 43:229–235
Zou Z, Zhao YG, Zhang L, **ao YH, Guo AP (2022) Analysis of Cyperus esculentus SMP family genes reveals lineage-specific evolution and seed desiccation-like transcript accumulation during tuber maturation. Ind Crop Prod 187:115382. https://doi.org/10.1016/j.indcrop.2022.115382
Acknowledgements
This work was supported by the Natural Science Foundation of Hainan province (320RC705), China; the Project of Sanya Yazhou Bay Science and Technology City (SCKJ-JYRC-2022-66), China; the Central Public-interest Scientific Institution Basal Research Fund (1630052022001), China; and the National Natural Science Foundation of China (31971688), China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Authors do not have any conflict of interest.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zou, Z., **ao, Y., Zhang, L. et al. Analysis of Lhc family genes reveals development regulation and diurnal fluctuation expression patterns in Cyperus esculentus, a Cyperaceae plant. Planta 257, 59 (2023). https://doi.org/10.1007/s00425-023-04092-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04092-5