Abstract
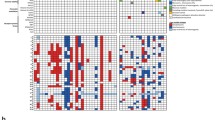
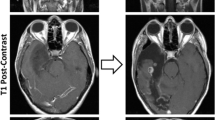
High-grade astrocytoma with piloid features (HGAP) is a recently recognized glioma type whose classification is dependent on its global epigenetic signature. HGAP is characterized by alterations in the mitogen-activated protein kinase (MAPK) pathway, often co-occurring with CDKN2A/B homozygous deletion and/or ATRX mutation. Experience with HGAP is limited and to better understand this tumor type, we evaluated an expanded cohort of patients (n = 144) with these tumors, as defined by DNA methylation array testing, with a subset additionally evaluated by next-generation sequencing (NGS). Among evaluable cases, we confirmed the high prevalence CDKN2A/B homozygous deletion, and/or ATRX mutations/loss in this tumor type, along with a subset showing NF1 alterations. Five of 93 (5.4%) cases sequenced harbored TP53 mutations and RNA fusion analysis identified a single tumor containing an NTRK2 gene fusion, neither of which have been previously reported in HGAP. Clustering analysis revealed the presence of three distinct HGAP subtypes (or groups = g) based on whole-genome DNA methylation patterns, which we provisionally designated as gNF1 (n = 18), g1 (n = 72), and g2 (n = 54) (median ages 43.5 years, 47 years, and 32 years, respectively). Subtype gNF1 is notable for enrichment with patients with Neurofibromatosis Type 1 (33.3%, p = 0.0008), confinement to the posterior fossa, hypermethylation in the NF1 enhancer region, a trend towards decreased progression-free survival (p = 0.0579), RNA processing pathway dysregulation, and elevated non-neoplastic glia and neuron cell content (p < 0.0001 and p < 0.0001, respectively). Overall, our expanded cohort broadens the genetic, epigenetic, and clinical phenotype of HGAP and provides evidence for distinct epigenetic subtypes in this tumor type.






Similar content being viewed by others
References
Alexander BM, Trippa L, Gaffey S, Arrillaga-Romany IC, Lee EQ, Rinne ML et al (2019) Individualized Screening Trial of Innovative Glioblastoma Therapy (INSIGhT): a Bayesian adaptive platform trial to develop precision medicines for patients with glioblastoma. JCO Precis Oncol. https://doi.org/10.1200/PO.18.00071
Bates D, Machler M, Bolker B, Walker S (2015) Fitting linear mixe-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Bender K, Perez E, Chirica M, Onken J, Kahn J, Brenner W et al (2021) High-grade astrocytoma with piloid features (HGAP): the Charite experience with a new central nervous system tumor entity. J Neurooncol 153:109–120. https://doi.org/10.1007/s11060-021-03749-z
Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D et al (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. https://doi.org/10.1038/nature26000
Cimino PJ, McFerrin L, Wirsching HG, Arora S, Bolouri H, Rabadan R et al (2018) Copy number profiling across glioblastoma populations has implications for clinical trial design. Neuro Oncol 20:1368–1373. https://doi.org/10.1093/neuonc/noy108
DeSisto J, Lucas JT Jr, Xu K, Donson A, Lin T, Sanford B et al (2021) Comprehensive molecular characterization of pediatric radiation-induced high-grade glioma. Nat Commun 12:5531. https://doi.org/10.1038/s41467-021-25709-x
Dodgshun AJ, Fukuoka K, Edwards M, Bianchi VJ, Das A, Sexton-Oates A et al (2020) Germline-driven replication repair-deficient high-grade gliomas exhibit unique hypomethylation patterns. Acta Neuropathol 140:765–776. https://doi.org/10.1007/s00401-020-02209-8
Ferreira R, Schneekloth JS Jr, Panov KI, Hannan KM, Hannan RD (2020) Targeting the RNA polymerase I transcription for cancer therapy comes of age. Cells. https://doi.org/10.3390/cells9020266
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. https://doi.org/10.1093/bioinformatics/btw313
Gutmann DH, McLellan MD, Hussain I, Wallis JW, Fulton LL, Fulton RS et al (2013) Somatic neurofibromatosis type 1 (NF1) inactivation characterizes NF1-associated pilocytic astrocytoma. Genome Res 23:431–439. https://doi.org/10.1101/gr.142604.112
Kessler T, Berberich A, Sadik A, Sahm F, Gorlia T, Meisner C et al (2020) Methylome analyses of three glioblastoma cohorts reveal chemotherapy sensitivity markers within DDR genes. Cancer Med 9:8373–8385. https://doi.org/10.1002/cam4.3447
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D et al (2021) The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. https://doi.org/10.1093/neuonc/noab106
Lucas CG, Sloan EA, Gupta R, Wu J, Pratt D, Vasudevan HN et al (2022) Multiplatform molecular analyses refine classification of gliomas arising in patients with neurofibromatosis type 1. Acta Neuropathol. https://doi.org/10.1007/s00401-022-02478-5
Mackay A, Burford A, Molinari V, Jones DTW, Izquierdo E, Brouwer-Visser J et al (2018) Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY Phase II Randomized Trial. Cancer Cell 33(829–842):e825. https://doi.org/10.1016/j.ccell.2018.04.004
Min JL, Hemani G, Davey Smith G, Relton C, Suderman M (2018) Meffil: efficient normalization and analysis of very large DNA methylation datasets. Bioinformatics 34:3983–3989. https://doi.org/10.1093/bioinformatics/bty476
Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F et al (2015) Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell 27:728–743. https://doi.org/10.1016/j.ccell.2015.04.002
Parker KA, Robinson NJ, Schiemann WP (2022) The role of RNA processing and regulation in metastatic dormancy. Semin Cancer Biol 78:23–34. https://doi.org/10.1016/j.semcancer.2021.03.020
Pratt D, Abdullaev Z, Papanicolau-Sengos A, Ketchum C, Panneer Selvam P, Chung HJ et al (2022) High-grade glioma with pleomorphic and pseudopapillary features (HPAP): a proposed type of circumscribed glioma in adults harboring frequent TP53 mutations and recurrent monosomy 13. Acta Neuropathol 143:403–414. https://doi.org/10.1007/s00401-022-02404-9
Pratt D, Lucas CG, Selvam PP, Abdullaev Z, Ketchum C, Quezado M et al (2022) Recurrent ACVR1 mutations in posterior fossa ependymoma. Acta Neuropathol 144:373–376. https://doi.org/10.1007/s00401-022-02435-2
Raffeld M, Abdullaev Z, Pack SD, ** L, Nagaraj S, Briceno N et al (2020) High level MYCN amplification and distinct methylation signature define an aggressive subtype of spinal cord ependymoma. Acta Neuropathol Commun 8:101. https://doi.org/10.1186/s40478-020-00973-y
Reinhardt A, Stichel D, Schrimpf D, Koelsche C, Wefers AK, Ebrahimi A et al (2019) Tumors diagnosed as cerebellar glioblastoma comprise distinct molecular entities. Acta Neuropathol Commun 7:163. https://doi.org/10.1186/s40478-019-0801-8
Reinhardt A, Stichel D, Schrimpf D, Sahm F, Korshunov A, Reuss DE et al (2018) Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol 136:273–291. https://doi.org/10.1007/s00401-018-1837-8
Ren X, Kuan PF (2019) methylGSA: a bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics 35:1958–1959. https://doi.org/10.1093/bioinformatics/bty892
Robinson JT, Thorvaldsdottir H, Wenger AM, Zehir A, Mesirov JP (2017) Variant review with the integrative genomics viewer. Cancer Res 77:e31–e34. https://doi.org/10.1158/0008-5472.CAN-17-0337
Singh O, Pratt D, Aldape K (2021) Immune cell deconvolution of bulk DNA methylation data reveals an association with methylation class, key somatic alterations, and cell state in glial/glioneuronal tumors. Acta Neuropathol Commun 9:148. https://doi.org/10.1186/s40478-021-01249-9
Stanley RF, Abdel-Wahab O (2022) Dysregulation and therapeutic targeting of RNA splicing in cancer. Nat Cancer 3:536–546. https://doi.org/10.1038/s43018-022-00384-z
Sturm D, Orr BA, Toprak UH, Hovestadt V, Jones DTW, Capper D et al (2016) New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164:1060–1072. https://doi.org/10.1016/j.cell.2016.01.015
Sturm D, Witt H, Hovestadt V, Khuong-Quang DA, Jones DT, Konermann C et al (2012) Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 22:425–437. https://doi.org/10.1016/j.ccr.2012.08.024
Tanguturi SK, Trippa L, Ramkissoon SH, Pelton K, Knoff D, Sandak D et al (2017) Leveraging molecular datasets for biomarker-based clinical trial design in glioblastoma. Neuro Oncol 19:908–917. https://doi.org/10.1093/neuonc/now312
Thomas C, Sill M, Ruland V, Witten A, Hartung S, Kordes U et al (2016) Methylation profiling of choroid plexus tumors reveals 3 clinically distinct subgroups. Neuro Oncol 18:790–796. https://doi.org/10.1093/neuonc/nov322
Yang B, Wang JQ, Tan Y, Yuan R, Chen ZS, Zou C (2021) RNA methylation and cancer treatment. Pharmacol Res 174:105937. https://doi.org/10.1016/j.phrs.2021.105937
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster.
Funding
This work was supported by the Intramural Research Program at the Center for Cancer Research, National Cancer Institute and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cimino, P.J., Ketchum, C., Turakulov, R. et al. Expanded analysis of high-grade astrocytoma with piloid features identifies an epigenetically and clinically distinct subtype associated with neurofibromatosis type 1. Acta Neuropathol 145, 71–82 (2023). https://doi.org/10.1007/s00401-022-02513-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-022-02513-5