Abstract
Objectives
To determine reproducible MRI parameters predictive of molecular subtype and risk stratification in glioma and develop a structured reporting system.
Methods
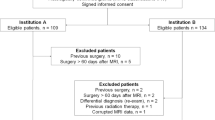
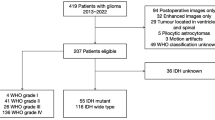
All study patients were initially diagnosed with glioma, 141 from the Cancer Genome Atlas and 131 from our tertiary institution, as training and validation sets, respectively. Images were analyzed by three neuroradiologists with 1–7 years of experience. MRI features including contrast enhancement pattern, necrosis, margin, edema, T2/FLAIR mismatch, internal cyst, and cerebral blood volume higher than normal cortex were reported using a structured reporting system. The pathology was stratified into five risk types: (1) oligodendroglioma, isocitrate dehydrogenase [IDH]-mutant, 1p19q co-deleted; (2) diffuse astrocytoma, IDH-mutant, grade II–III; (3) glioblastoma, IDH-mutant, grade IV; (4) diffuse astrocytoma, IDH-wild, grade II–III; and (5) glioblastoma, IDH-wild, grade IV. Significant predictors were selected using multivariate logistic regression, and diagnostic performance was tested using a validation set.
Results
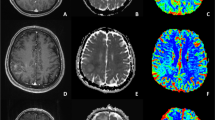
Reproducible imaging parameters exhibiting > 50% agreement across readers included the presence of necrosis, T2/FLAIR mismatch, internal cyst, and predominant contrast enhancement. In the validation set, prediction of risk type 5 exhibited the highest diagnostic performance with AUCs of 0.92 (reader 1) and 0.93 (reader 2) with predominant enhancement, followed by risk type 2 with AUCs of 0.95 and 0.95 with T2/FLAIR mismatch sign and no necrosis, and risk type 1 with AUCs of 0.84 and 0.83 with internal cyst or necrosis. Risk types 3 and 4 were difficult to visually predict.
Conclusions
Imaging parameters with high reproducibility enabling prediction of IDH-wild-type glioblastoma, IDH-mutant/1p19q co-deletion oligodendroglioma, and IDH-mutant diffuse astrocytoma were identified.
Key Points
• Reproducible MRI parameters for determining molecular subtypes of glioma included the presence of necrosis, T2/FLAIR mismatch, internal cyst, and predominant contrast enhancement.
• IDH-wild type glioblastoma, IDH-mutant/1p19q co-deletion oligodendroglioma, and IDH-mutant low-grade astrocytoma were identified using MRI parameters with high inter-reader reproducibility.
• Identification of IDH-wild type low-grade glioma and IDH-mutant glioblastoma was difficult by visual analysis.



Similar content being viewed by others
Abbreviations
- 1p/19q:
-
Chromosome arms 1p and 19q
- CBV:
-
Cerebral blood volume
- FLAIR:
-
Fluid-attenuated inversion recovery
- IDH:
-
Isocitrate dehydrogenase
- TCGA:
-
The Cancer Genome Atlas
- TCIA:
-
The Cancer Imaging Archive
- VASARI:
-
Visually AcceSAble Rembrandt Images
- WHO:
-
World Health Organization
References
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Hu N, Richards R, Jensen R (2016) Role of chromosomal 1p/19q co-deletion on the prognosis of oligodendrogliomas: a systematic review and meta-analysis. Interdiscip Neurosurg 5:58–63
Yan H, Parsons DW, ** G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Broen MPG, Smits M, Wijnenga MMJ et al (2018) The T2-FLAIR mismatch sign as an imaging marker for non-enhancing IDH-mutant, 1p/19q-intact lower-grade glioma: a validation study. Neuro Oncol 20:1393–1399
American College of Radiology, D'Orsi CJ (2013) ACR BI-RADS Atlas: Breast Imaging Reporting and Data System; Mammography, Ultrasound, Magnetic Resonance Imaging, Follow-up and Outcome Monitoring, Data Dictionary. ACR, American College of Radiology. https://books.google.co.kr/books?id=AoWfnQAACAAJ
Zhou H, Vallieres M, Bai HX et al (2017) MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 19:862–870
Park Y, Han K, Ahn S et al (2018) Prediction of IDH1-mutation and 1p/19q-codeletion status using preoperative MR imaging phenotypes in lower grade gliomas. AJNR Am J Neuroradiol 39:37–42
Chen X, Fang M, Dong D et al (2019) Development and validation of a MRI-based radiomics prognostic classifier in patients with primary glioblastoma multiforme. Acad Radiol 26:1292–1300
Due-Tønnessen P, Pinho MC, Emblem KE et al (2019) The impact of MRI features and observer confidence on the treatment decision-making for patients with untreated glioma. Sci Rep 9:19898
Hyare H, Rice L, Thust S et al (2019) Modelling MR and clinical features in grade II/III astrocytomas to predict IDH mutation status. Eur J Radiol 114:120–127
Ivanidze J, Lum M, Pisapia D et al (2019) MRI features associated with TERT promoter mutation status in glioblastoma. J Neuroimaging 29:357–363
Lee M, Han K, Ahn SS et al (2019) The added prognostic value of radiological phenotype combined with clinical features and molecular subtype in anaplastic gliomas. J Neurooncol 142:129–138
Peeken JC, Goldberg T, Pyka T et al (2019) Combining multimodal imaging and treatment features improves machine learning-based prognostic assessment in patients with glioblastoma multiforme. Cancer Med 8:128–136
Su CQ, Lu SS, Han QY et al (2019) Intergrating conventional MRI, texture analysis of dynamic contrast-enhanced MRI, and susceptibility weighted imaging for glioma grading. Acta Radiol 60:777–787
Su CQ, Lu SS, Zhou MD et al (2019) Combined texture analysis of diffusion-weighted imaging with conventional MRI for non-invasive assessment of IDH1 mutation in anaplastic gliomas. Clin Radiol 74:154–160
Lasocki A, Tsui A, Gaillard F et al (2017) Reliability of noncontrast-enhancing tumor as a biomarker of IDH1 mutation status in glioblastoma. J Clin Neurosci 39:170–175
Clark K, Vendt B, Smith K et al (2013) The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26:1045–1057
Leu K, Ott GA, Lai A et al (2017) Perfusion and diffusion MRI signatures in histologic and genetic subtypes of WHO grade II–III diffuse gliomas. J Neurooncol 134:177–188
Eckel-Passow JE, Lachance DH, Molinaro AM et al (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508
Kanazawa T, Fujiwara H, Takahashi H et al (2019) Imaging scoring systems for preoperative molecular diagnoses of lower-grade gliomas. Neurosurg Rev 42:433–441
Maynard J, Okuchi S, Wastling S et al (2020) World Health Organization grade II/III glioma molecular status: prediction by MRI morphologic features and apparent diffusion coefficient. Radiology 296:111–121
Niemeyer B, Muniz B, Marchiori E (2018) T2-FLAIR mismatch sign as an imaging biomarker in lower-grade gliomas. Eur Neurol 79:317–318
Wang X (2014) Firth logistic regression for rare variant association tests. Front Genet 5:187
Rudie JD, Rauschecker AM, Bryan RN et al (2019) Emerging applications of artificial intelligence in neuro-oncology. Radiology 290:607–618
Carrillo J, Lai A, Nghiemphu P et al (2012) Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol 33:1349–1355
Choi C, Ganji SK, DeBerardinis RJ et al (2012) 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 18:624–629
Koeller KK, Rushing EJ (2005) Oligodendroglioma and its variants: radiologic-pathologic correlation. Radiographics 25:1669–1688
Zhao S-S, Feng X-L, Hu Y-C et al (2020) Better efficacy in differentiating WHO grade II from III oligodendrogliomas with machine-learning than radiologist’s reading from conventional T1 contrast-enhanced and fluid attenuated inversion recovery images. BMC Neurol 20:1–10
Bankier AA, Levine D, Halpern EF et al (2010) Consensus interpretation in imaging research: is there a better way? Radiology 257:14–17
Molinaro AM, Taylor JW, Wiencke JK et al (2019) Genetic and molecular epidemiology of adult diffuse glioma. Nat Rev Neurol 15:405–417
Park M, Kim H, Jahng G-H et al (2009) Semiquantitative assessment of intratumoral susceptibility signals using non-contrast-enhanced high-field high-resolution susceptibility-weighted imaging in patients with gliomas: comparison with MR perfusion imaging. AJNR Am J Neuroradiol 30:1402–1408
Gevaert O, Mitchell LA, Achrol AS et al (2014) Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 273:168–174
Yu J, Shi Z, Lian Y et al (2017) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 27:3509–3522
Eichinger P, Alberts E, Delbridge C et al (2017) Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas. Sci Rep 7:13396
Chang K, Bai HX, Zhou H et al (2018) Residual convolutional neural network for the determination of IDH status in low- and high-grade gliomas from MR imaging. Clin Cancer Res 24:1073–1081
Chang P, Grinband J, Weinberg BD et al (2018) Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas. AJNR Am J Neuroradiol 39:1201–1207
Han L, Kamdar MR (2018) MRI to MGMT: predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. Pac Symp Biocomput 23:331–342
Liang S, Zhang R, Liang D et al (2018) Multimodal 3D DenseNet for IDH genotype prediction in gliomas. Genes (Basel) 9(8):382
Funding
This research was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP) (grant numbers: NRF-2020R1A2B5B01001707 and NRF-2020R1A2C4001748).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Ho Sung Kim.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors has significant statistical expertise (Seo Young Park, 8 years of experienced statistician).
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• cross-sectional study
• performed at different institutions
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 8896 kb)
Rights and permissions
About this article
Cite this article
Nam, Y.K., Park, J.E., Park, S.Y. et al. Reproducible imaging-based prediction of molecular subtype and risk stratification of gliomas across different experience levels using a structured reporting system. Eur Radiol 31, 7374–7385 (2021). https://doi.org/10.1007/s00330-021-08015-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-021-08015-4