Abstract
Objectives
To assess the combined role of tumor vascularity, estimated from perfusion MRI, and MGMT methylation status on overall survival (OS) in patients with glioblastoma.
Methods
A multicentric international dataset including 96 patients from NCT03439332 clinical study were used to study the prognostic relationships between MGMT and perfusion markers. Relative cerebral blood volume (rCBV) in the most vascularized tumor regions was automatically obtained from preoperative MRIs using ONCOhabitats online analysis service. Cox survival regression models and stratification strategies were conducted to define a subpopulation that is particularly favored by MGMT methylation in terms of OS.
Results
rCBV distributions did not differ significantly (p > 0.05) in the methylated and the non-methylated subpopulations. In patients with moderately vascularized tumors (rCBV < 10.73), MGMT methylation was a positive predictive factor for OS (HR = 2.73, p = 0.003, AUC = 0.70). In patients with highly vascularized tumors (rCBV > 10.73), however, there was no significant effect of MGMT methylation (HR = 1.72, p = 0.10, AUC = 0.56).
Conclusions
Our results indicate the existence of complementary prognostic information provided by MGMT methylation and rCBV. Perfusion markers could identify a subpopulation of patients who will benefit the most from MGMT methylation. Not considering this information may lead to bias in the interpretation of clinical studies.
Key Points
• MRI perfusion provides complementary prognostic information to MGMT methylation.
• MGMT methylation improves prognosis in glioblastoma patients with moderate vascular profile.
• Failure to consider these relations may lead to bias in the interpretation of clinical studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas are the most common malignant primary central nervous system tumors, with an estimated annual incidence of 3.21 per 100,000 individuals in the USA [1]. About half of all newly diagnosed gliomas are classified as glioblastoma, which is the most malignant type of brain cancer. Glioblastoma pathology is characterized by angiogenesis, highly infiltrative growth, and cellular heterogeneity [2]. Despite an aggressive therapeutic approach combining maximum safe resection with radiotherapy (RT) plus concomitant and adjuvant temozolomide (TMZ), prognosis is poor, with median overall survival (OS) duration of approximately 14 months [3]. In 2016, the World Health Organization introduced molecular parameters along with histology to describe the interpatient glioblastoma heterogeneity associated with differential prognosis and responses to therapy [4].
Adequate clinical and molecular biomarkers are needed for accurate estimations of prognosis and optimal treatment selections. Inactivation through promoter methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene, which impairs the ability to repair DNA damaged induced by alkylating agents such as TMZ [5], has been described as a relevant biomarker for clinical decision-making in glioblastoma treatment. In a post hoc analysis of a phase III trial, MGMT promoter methylation was associated with a 2-year survival increase in TMZ-treated glioblastoma patients from 14 to 46% [6]. Additional studies have also described not only a predictive, but also a prognostic role of MGMT methylation for glioblastoma patients [7, 8]. Current guidelines support the use of MGMT methylation as a predictive biomarker in patients older than 70 years with isocitrate dehydrogenase (IDH) wild-type grade IV gliomas [8].
Despite the well-documented impact of MGMT methylation on the prognosis of glioblastoma patients treated with TMZ, the survival of these patients is not explained by this factor alone. Tumor vascularity, for instance, is strongly associated with glioma transformation (i.e. grade progression), poorer survival [9], sensitivity to RT, and effectiveness of bloodborne delivery of nutrients and chemotherapy [10].
MRI perfusion–based parameters correlate with tumor vascularity and properties of vessels [11, 12]. Numerous studies show that the vascularity as defined by MRI perfusion is a prognostic factor for glioblastoma even prior to initial surgery [13,14,15]. Perfusion parameters, such as relative cerebral blood volume (rCBV) in the enhancing tumor areas, are among the most consistently recognized independent predictors of survival [9]. Novel approaches based on artificial intelligence [15, 16] have been proposed to calculate perfusion-based biomarkers, based not on the entire enhancing lesion but on more homogeneous regions (habitats), thereby improving not only the prognostic capacity but also the reproducibility of the results [13, 17, 18].
In this study, we aimed at evaluating whether the rCBV is a modulating factor of the prognostic effect of MGMT methylation status in patients with glioblastoma treated with TMZ.
Materials and methods
Patient cohort
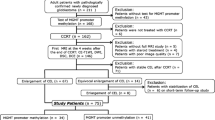
The patient cohort has compiled by the multicenter and international retrospective clinical study NCT03439332 [19]. A total of 110 cases from this dataset with MRI perfusion studies were included in the current study (Fig. 1). The NCT03439332 multicenter international dataset included patients treated at seven European hospitals from four countries: Hospital Clinic, Barcelona, Spain; Hospital Universitario Vall d’Hebron, Barcelona, Spain; Hospital Universitario de La Ribera, Alzira, Spain; Hospital de Manises, Manises, Spain; Azienda Ospedaliero-Universitaria di Parma, Parma, Italy; Oslo University Hospital, Oslo, Norway; and Centre Hospitalier Universitaire de Liège, Liège, Belgium. Patients were diagnosed with glioblastoma grade IV WHO with histopathological confirmation and followed Stupp standard treatment. The extent of resection was assessed in each center by expert neurosurgeons and radiologists based on the postsurgical MRI study findings. A material transfer agreement was approved by all the participating centers and an acceptance report was issued by the Ethical Committee of each center. The Universitat Politècnica de València institutional ethical board also approved this retrospective study. All methods were performed in accordance with the relevant guidelines and regulations or Declaration of Helsinki.
IDH1 mutation and MGMT methylation status assessment
MGMT methylation status was assessed in each center by pyrosequencing using a cut-off value between 9 and 10%, except for the cases from the Hospital Clinic in Barcelona where MGMT methylation status was assessed by methylation-specific polymerase chain reaction. The IDH1 mutation status was assessed in each center by immunostains using the IDH1 R132H antibody.
MRI data acquisition protocol
Pre-surgical standard-of-care MRI examinations including pre- and post-gadolinium T1-weighted MRI as well as T2-weighted, T2-fluid-attenuated inversion recovery (FLAIR), and dynamic susceptibility contrast (DSC) T2* perfusion-weighted sequences were collected from each participating center. All the MRIs were obtained by 1.5-T or 3-T scanners, using different MRI acquisition protocols in each participating center. A summary of the MRI acquisition protocol used by each center is presented in Supplementary Materials Table S-I.
MRI data preprocessing
Anatomical MRI acquisitions were preprocessed using the following pipeline: (1) voxel isotropic resampling, (2) denoising, (3) rigid intra-patient registration, (4) affine registration to MNI space [20], (5) brain extraction, and (6) magnetic field inhomogeneity correction. First, image resampling at 1 mm3 was performed through linear interpolation. Next, denoising was carried out using the adaptive non-local means filter [21] with a search window of 7 × 7 × 7 voxels and a patch window of 3 × 3 × 3 voxels. Registration was performed with ANTs software [22] and Mutual Information, using the T1c MRI as reference. Brain extraction was performed through a convolutional neural network with a U-Net architecture trained on a dataset of 160 T1c MRIs with glioblastomas manually annotated and validated by several independent neuroimaging researchers with more than 5 years of experience. Finally, magnetic field inhomogeneities were corrected with the N4 software using the previously computed intra-cranial mask [23].
Lesion segmentation
Glioblastoma tissue segmentation was performed by means of 3D patch-based convolutional neural networks [24]. A U-Net Res-Net architecture of 5 levels with long-term skip connections with 16, 32, 64, 128, and 256 filters at each level, respectively, was used to perform the segmentation. At each level, a simple block and a residual block were chained together. A simple block consists of a convolution + batch normalization + ReLu activation function. A residual block consists of a convolution + batch normalization + ReLu + convolution + batch normalization + residual connection summation + ReLu activation function. Convolutions were performed with isotropic kernels of size 3 × 3 × 3, while batch normalization momentum was fixed to 0.9. Max-pooling layers with pooling size and stride 2 × 2 × 2 were employed in the contracting path to sequentially condensate the relevant features of the input patches, while transpose convolutions were used in the expanding path to rearrange and project the latent features to the original size. The network works with patches of 32 × 32 × 32 with 3 channels corresponding to the T1c, T2, and FLAIR sequences. Adam optimizer with cross-entropy loss-function was used to train the network. The lesion segmentation network was trained on BraTS dataset [25,26,32].
As suggested in the literature [6], we performed a Kaplan-Meier survival analysis to assess differences in OS between patient subpopulations. The log-rank test was performed to test for significant differences in OS. For censored cases, we set the date of censorship to the last date of contact with the patient or, in cases where this information was not available, the date of the last MRI exam.
Finally, to reduce biases in survival analyses, we analyzed whether the IDH1 mutation was related to MGMT methylation by performing a Fisher test, and whether IDH1-mutated tumors have significantly different rCBVHAT values performing a Mann-Whitney U test, using those cases where IDH1 information was available.
Software
The MRI data analysis was performed using ONCOhabitats [24], a freely available online service (www.onchabitats.upv.es). It provides a fully automated pipeline for the analysis of glioblastoma MRI studies including pre-processing, lesion segmentation, DSC perfusion quantification, and high angiogenic tumor delineation services, among others. Statistical analysis was performed with MATLAB (R2019a) and R (v3.6.0) [33].
Results
Characterization of patient cohorts
Of the 110 patients included from the NCT03439332 dataset, six exams (patients) were excluded because of processing errors with the automatic processing pipeline, five exams were excluded due to noise or MR artifacts that precluded DSC quantification (gamma-variate R2 goodness of fit < 0.95), one exam was excluded due to inability to differentiate between tumor vascularity and reactive meningeal enhancement, and two cases were excluded because MRI perfusion field of view did not cover the entire lesion (see Fig. 1). Of the remaining 96 patients, 43 (44.8%) presented the MGMT methylated. With a median follow-up of 391 days, median OS was 402 days; 15 patients were alive at the last data cut-off date and were censored in the survival analyses. Table 1 includes the most relevant demographic, molecular, and clinical features of the study cohort and summarizes the patients recruited. No significant relationships have been found between IDH1 mutation and MGMT methylation status (p = 0.39), nor between IDH1 mutation and rCBVHAT values (p = 0.15).
Non-significant association between MGMT and rCBVHAT
Median rCBVHAT in the whole population was 10.73. Median rCBVHAT values in methylated and non-methylated MGMT subpopulations were 11.06 and 10.35, respectively. rCBVHAT distributions did not differ significantly (Mann-Whitney U of 1062; n = 43 and n = 53, respectively, p > 0.05 two-tailed) in the methylated and the non-methylated subpopulations.
MGMT methylation status and rCBVHAT provide complementary prognostic information
In order to evaluate the association between rCBVHAT (as a continuous variable) and MGMT methylation status with OS, we initially constructed three Cox proportional hazards (Cox-PH) regression models. The results of the three Cox-PH regression models obtained from NCT03439332 dataset are shown in Table 2. Results of the two first models including rCBVHAT or MGMT status independently showed a significant association of MGMT status (HR: 2.01; 95% CI: 1.21–3.34; p = 0.007) with OS, but a non-significant association of rCBVHAT (HR: 1.05; 95% CI: 0.99–1.11; p = 0.102) with OS. When evaluating both parameters in the multivariable Cox-PH model (Table 2), both were independently associated with OS: MGMT status (HR: 2.12; 95% CI: 1.27–3.52; p = 0.004) and rCBVHAT (HR: 1.06; 95% CI: 1.00–1.12; p = 0.049). The performance of the model improved by increasing the concordance, decreasing the p values of each variable, and increasing the hazard ratios (HR), suggesting that MGMT methylation and rCBVHAT provide complementary prognostic information.
rCBVHAT improves the capability of MGMT to stratify responsive patients
We then evaluated the impact of MGMT methylation status in patients with highly and moderately vascularized tumors, as defined by the rCBVHAT values. rCBVth, defined as the median rCBVHAT of the study cohort, was established at rCBVth = 10.73.
We then analyzed differences in OS between patients with methylated and unmethylated MGMT promoters for the entire cohort, patients with highly vascularized tumors (rCBVHAT ≥ 10.73), and patients with moderately vascularized tumors (rCBVHAT < 10.73). In the cohort, 48 (50%) and 48 (50%) patients presented with highly and moderately vascularized tumors, respectively.
We observed a significant association of MGMT methylation and OS (HR: 2.1 [95% CI: 1.33–3.33]; p = 0.002) (Table 3). In patients with moderately vascularized tumors (rCBVHAT < 10.73), the association between MGMT methylation and OS was higher (HR: 2.73 [95% CI: 1.40–5.32]; p = 0.003) than in the general population. Finally, there was no association between MGMT methylation and OS in patients with high vascularity (rCBVHAT ≥ 10.73) (HR: 1.72 [95% CI: 0.90–3.27]; p = 0.10). Kaplan-Meier curves for the four subpopulations are presented in Fig. 3.
Discussion
In this study, we present a potential role of tumor vascularity in the prognostic impact of MGMT methylation status in glioblastoma patients. As was expected from the literature, we observe a significant prognostic impact of MGMT methylation in glioblastoma patients treated with TMZ included in the NCT03439332 multicenter international dataset. Moreover, our results show that tumor vascularity may identify a subgroup of patients that exhibit a greater benefit from MGMT methylation. Once glioblastoma patients are dichotomized into highly and moderately vascularized tumor subgroups using rCBV, we observe a highly significant impact of MGMT status in patients with moderately vascularized tumors and only a non-significant trend in patients with highly vascularized tumors. From the comparison of the three multiparametric Cox survival analyses based on MGMT and rCBVHAT, we conclude that the information provided by both variables are relevant and complementary in predicting the prognosis of glioblastoma patients treated with TMZ. This implies that MGMT methylation assessment may provide incomplete information regarding prognosis, and that adding the characterization of tumor vascularization may improve estimation of responsiveness to TMZ. We propose a new classification of patients based on tumor vascularity, derived from the NCT03439332 multicenter international dataset. Our study proves the feasibility of using perfusion MRIs to identify subpopulations of glioblastoma patients with a higher likelihood of a beneficial effect of standard Stupp treatment (i.e., including TMZ), patients with methylated MGMT, and moderate vascularization in the contrast-enhancing tumor areas. On the contrary, patients with highly vascularized tumors will probably benefit less from TMZ, independently of the MGMT methylation status. We hypothesize that tumors with a lower vascularization may be potentially less aggressive, with a lower prevalence of molecular aberrations that may confer resistance to alkylating agents in the presence of a methylated MGMT.
Different studies have evaluated possible associations between MGMT methylation and tumor vascularity. A study by Hempel et al [34] (n = 100 including 24 glioblastomas) found significantly higher rCBV values in IDH wild-type glioblastomas with a methylated MGMT than in those with an unmethylated MGMT. Chahal et al [35] found higher levels of vascular endothelial growth factor receptor 1 (VEGFR-1) in unmethylated compared with methylated MGMT cells, hypothesizing that this should lead to an increased vascularization of the tumor. However, another small sample study (n = 24) by Moon et al [36] concluded that rCBV did not differ significantly between methylated and unmethylated MGMT tumors. In our study, based on a larger cohort, no significant relationship was observed between MGMT methylation and rCBVHAT. Due to the influence of MGMT on the effectiveness of therapies with antineoplastic drugs of alkylating agent class (i.e., TMZ), several studies suggest avoiding these therapies in those patients with non-methylated MGMT [6, 37]. The conclusions of this study further suggest that to obtain the most significant benefit, we should select among patients with methylated MGMT those with moderate vascularity.
The main limitation of the study is the lack of a complete molecular profile for all cases. Several studies already showed the importance of combining MGMT methylation status with other features when determining the patients who will benefit the most from it. Having methylated MGMT is an advantage especially when TERT expression is high [38] and progression-free survival is higher when both MGMT and mutant p53 expression are low [39]. Also, MGMT methylation displays better survival prediction performance when combined with IDH1 mutations [40]. In this study, we have chosen to use CBV as the most relevant perfusion parameter for the analysis of the prognostic value of vascularity. However, in future work, it would be interesting to extend the study to include complementary markers derived from morphological [41], molecular [42], dynamic contrast-enhanced, and vessel architecture [43] imaging sequences that could help us to understand the complex vascular phenomena behind the results of this study.
This study demonstrates that combining MGMT methylation status together with the rCBVHAT value obtained completely automatically from standard-of-care MRI studies through an online and open image analysis service (https://www.oncohabitats.upv.es) can provide a more reliable prognostic indicator for designing patient stratification strategies. This ensures the reproducibility of the obtained results and the immediate applicability of the proposed conclusions in the clinical trial setting. In conclusion, we consider that information of rCBVHAT and MGMT methylation status should be included in randomization/stratification strategies of new clinical trials. Due to the joint implications of these two factors on patient survival, failure to consider these factors may lead to significant bias in the interpretation of the results from such studies.
Abbreviations
- AUC:
-
Area under the curve
- DSC:
-
Dynamic susceptibility contrast
- FLAIR:
-
Fluid-attenuated inversion recovery
- HAT:
-
Highly angiogenic tumor
- HR :
-
Hazard ratio
- IDH:
-
Isocitrate dehydrogenase
- KPS:
-
Karnofsky Performance Status
- MGMT :
-
O(6)-Methylguanine-DNA methyltransferase
- MS-MLPA:
-
Methylation-specific multiplex ligation-dependent probe amplification
- MSP:
-
Methylation-specific polymerase chain reaction
- OS:
-
Overall survival
- rCBF:
-
Relative cerebral blood flow
- rCBV:
-
Relative cerebral blood volume
- RT:
-
Radiotherapy
- TMZ:
-
Temozolomide
References
Ostrom QT, Gittleman H, Liao P et al (2017) CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro Oncol 19:v1–v88. https://doi.org/10.1093/neuonc/nox158
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507. https://doi.org/10.1056/NEJMra0708126
Van Meir EG, Hadjipanayis CG, Norden AD, Shu HK, Wen PY, Olson JJ (2010) Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 60:166–193. https://doi.org/10.3322/caac.20069
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Stupp R, Hegi ME, Mason WP et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10:459–466. https://doi.org/10.1016/S1470-2045(09)70025-7
Hegi ME, Diserens A-C, Gorlia T et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003. https://doi.org/10.1056/NEJMoa043331
Gorlia T, van den Bent MJ, Hegi ME et al (2008) Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol 9:29–38. https://doi.org/10.1016/S1470-2045(07)70384-4
Rivera AL, Pelloski CE, Gilbert MR et al (2010) MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol 12:116–121. https://doi.org/10.1093/neuonc/nop020
Russell SM, Elliott R, Forshaw D, Golfinos JG, Nelson PK, Kelly PJ (2009) Glioma vascularity correlates with reduced patient survival and increased malignancy. Surg Neurol 72:242–246; discussion 246-247. https://doi.org/10.1016/j.surneu.2008.11.012
Batchelor TT, Gerstner ER, Emblem KE et al (2013) Improved tumor oxygenation and survival in glioblastoma patients who show increased blood perfusion after cediranib and chemoradiation. Proc Natl Acad Sci U S A 110:19059–19064. https://doi.org/10.1073/pnas.1318022110
Ulyte A, Katsaros VK, Liouta E et al (2016) Prognostic value of preoperative dynamic contrast-enhanced MRI perfusion parameters for high-grade glioma patients. Neuroradiology 58:1197–1208. https://doi.org/10.1007/s00234-016-1741-7
Yoo R-E, Yun TJ, Hwang I et al (2020) Arterial spin labeling perfusion-weighted imaging aids in prediction of molecular biomarkers and survival in glioblastomas. Eur Radiol 30:1202–1211. https://doi.org/10.1007/s00330-019-06379-2
Fuster-Garcia E, Juan-Albarracín J, García-Ferrando GA et al (2018) Improving the estimation of prognosis for glioblastoma patients by MR based hemodynamic tissue signatures. NMR Biomed 31:e4006. https://doi.org/10.1002/nbm.4006
Hou BL, Wen S, Katsevman GA et al (2018) Magnetic resonance imaging parameters and their impact on survival of patients with glioblastoma: tumor perfusion predicts survival. World Neurosurg. https://doi.org/10.1016/j.wneu.2018.12.085
Juan-Albarracín J, Fuster-Garcia E, Pérez-Girbés A et al (2018) Glioblastoma: vascular habitats detected at preoperative dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging predict survival. Radiology 287:944–954. https://doi.org/10.1148/radiol.2017170845
Chang Y-CC, Ackerstaff E, Tschudi Y et al (2017) Delineation of tumor habitats based on dynamic contrast enhanced MRI. Sci Rep 7:9746. https://doi.org/10.1038/s41598-017-09932-5
Álvarez-Torres MDM, Juan-Albarracín J, Fuster-Garcia E et al (2019) Robust association between vascular habitats and patient prognosis in glioblastoma: an international multicenter study. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26958
Wu H, Tong H, Du X et al (2020) Vascular habitat analysis based on dynamic susceptibility contrast perfusion MRI predicts IDH mutation status and prognosis in high-grade gliomas. Eur Radiol. https://doi.org/10.1007/s00330-020-06702-2
Multicentre validation of how vascular biomarkers from tumor can predict the survival of the patient with glioblastoma - full text view - ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT03439332. Accessed 24 Apr 2019
Brett M, Johnsrude IS, Owen AM (2002) The problem of functional localization in the human brain. Nat Rev Neurosci 3:243–249. https://doi.org/10.1038/nrn756
Manjón JV, Coupé P, Martí-Bonmatí L, Collins DL, Robles M (2010) Adaptive non-local means denoising of MR images with spatially varying noise levels. J Magn Reson Imaging 31:192–203. https://doi.org/10.1002/jmri.22003
Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC (2011) A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54:2033–2044. https://doi.org/10.1016/j.neuroimage.2010.09.025
Tustison NJ, Avants BB, Cook PA et al (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320. https://doi.org/10.1109/TMI.2010.2046908
Juan-Albarracín J, Fuster-Garcia E, García-Ferrando GA, García-Gómez JM (2019) ONCOhabitats: a system for glioblastoma heterogeneity assessment through MRI. Int J Med Inform 128:53–61. https://doi.org/10.1016/j.ijmedinf.2019.05.002
Menze BH, Jakab A, Bauer S et al (2015) The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 34:1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Bakas S, Akbari H, Sotiras A et al (2017) Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4:170117. https://doi.org/10.1038/sdata.2017.117
Bakas S, Reyes M, Jakab A, et al (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. Ar**v181102629
Juan-Albarracín J, Fuster-Garcia E, del Mar Álvarez-Torres M et al (2020) ONCOhabitats glioma segmentation model. In: Crimi A, Bakas S (eds) Brain lesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Springer International Publishing, Cham, pp 295–303
Knutsson L, Ståhlberg F, Wirestam R (2010) Absolute quantification of perfusion using dynamic susceptibility contrast MRI: pitfalls and possibilities. MAGMA 23:1–21. https://doi.org/10.1007/s10334-009-0190-2
Boxerman JL, Schmainda KM, Weisskoff RM (2006) Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am J Neuroradiol 27:859–867
Little RJA, Rubin DB (2002) Statistical analysis with missing data. John Wiley & Sons, New York [etc.]
Harrell FE, Lee KL, Mark DB (1996) Multivariable prognostic models: issues in develo** models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 15:361–387. https://doi.org/10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4
R Core Team (v3.6.0) R: a language and environment for statistical computing
Hempel J-M, Schittenhelm J, Klose U et al (2018) In vivo molecular profiling of human glioma. Clin Neuroradiol. https://doi.org/10.1007/s00062-018-0676-2
Chahal M, Xu Y, Lesniak D et al (2010) MGMT modulates glioblastoma angiogenesis and response to the tyrosine kinase inhibitor sunitinib. Neuro Oncol 12:822–833. https://doi.org/10.1093/neuonc/noq017
Moon W-J, Choi JW, Roh HG, Lim SD, Koh YC (2012) Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology 54:555–563. https://doi.org/10.1007/s00234-011-0947-y
Li Q, Guo J, Wang W, Wang D (2017) Relationship between MGMT gene expression and treatment effectiveness and prognosis in glioma. Oncol Lett 14:229–233. https://doi.org/10.3892/ol.2017.6123
Nguyen HN, Lie A, Li T et al (2017) Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol 19:394–404. https://doi.org/10.1093/neuonc/now189
Li S, Jiang T, Li G, Wang Z (2008) Impact of p53 status to response of temozolomide in low MGMT expression glioblastomas: preliminary results. Neurol Res 30:567–570. https://doi.org/10.1179/174313208X297913
Molenaar RJ, Verbaan D, Lamba S et al (2014) The combination of IDH1 mutations and MGMT methylation status predicts survival in glioblastoma better than either IDH1 or MGMT alone. Neuro Oncol 16:1263–1273. https://doi.org/10.1093/neuonc/nou005
Li Z-C, Bai H, Sun Q et al (2018) Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol 28:3640–3650. https://doi.org/10.1007/s00330-017-5302-1
Jiang S, Rui Q, Wang Y et al (2018) Discriminating MGMT promoter methylation status in patients with glioblastoma employing amide proton transfer-weighted MRI metrics. Eur Radiol 28:2115–2123. https://doi.org/10.1007/s00330-017-5182-4
Emblem KE, Mouridsen K, Bjornerud A et al (2013) Vessel architectural imaging identifies cancer patient responders to anti-angiogenic therapy. Nat Med 19:1178–1183. https://doi.org/10.1038/nm.3289
Acknowledgments
We sincerely thank the collaboration of the different hospitals that have participated in the NCT03439332 multicenter international study: Hospital Clinic, Barcelona, Spain; Hospital Universitario Vall d’Hebron, Barcelona, Spain; Hospital Universitario de La Ribera, Alzira, Spain; Hospital de Manises, Manises, Spain; Azienda Ospedaliero-Universitaria di Parma, Parma, Italy; Oslo University Hospital, Oslo, Norway; and Centre Hospitalier Universitaire de Liège, Liège, Belgium.
Funding
Open Access funding provided by University of Oslo (incl Oslo University Hospital). This study has received funding from MTS4up project (National Plan for Scientific and Technical Research and Innovation 2013-2016, No. DPI2016-80054-R) (JMGG); H2020-SC1-2016-CNECT Project (No. 727560) (JMGG), H2020-SC1-BHC-2018-2020 (No. 825750) (JMGG), the European Research Council (ERC) under the European Union’s Horizon 2020 (Grant Agreement No. 758657), the South-Eastern Norway Regional Health Authority Grants 2017073 and 2013069, the Research Council of Norway Grants 261984 (KEE). M.A.T was supported by Programa Estatal de Promoción del Talento y su Empleabilidad en I+D+i (DPI2016-80054-R). E.F.G was supported by the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement (No. 844646).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Juan Miguel García Gómez, full professor at Universitat Politècnica de València (juanmig@upv.es).
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained. A material transfer agreement was approved by all the participating centers and an acceptance report was issued by the Ethical Committee of each center. The Universitat Politècnica de València institutional ethical board also approved this retrospective study.
Study subjects or cohorts overlap
Some study subjects or cohorts have been previously reported in [16].
In the present study, we used a subset of the NCT03439332 multicenter study cohort. The NCT03439332 cohort was used in [16] to validate the primary endpoint of the NCT03439332 study, which is to validate the robust association between vascular habitats and patient prognosis in glioblastoma. In the present study, however, we use a subset of these data to study the joint influence of vascularity, estimated by perfusion MRI, and MGMT methylation on the survival of patients with GBM. MGMT methylation was not considered in the study of Álvarez-Torres et al [16]; moreover, the objective, results, and conclusions do not overlap between the current and previous studies.
[16] Álvarez-Torres MDM, Juan-Albarracín J, Fuster-Garcia E, et al (2019) Robust association between vascular habitats and patient prognosis in glioblastoma: an international multicenter study. J Magn Reson Imaging. https://doi.org/10.1002/jmri.26958
Methodology
•retrospective
•observational
•multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 23 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fuster-Garcia, E., Lorente Estellés, D., Álvarez-Torres, M. et al. MGMT methylation may benefit overall survival in patients with moderately vascularized glioblastomas. Eur Radiol 31, 1738–1747 (2021). https://doi.org/10.1007/s00330-020-07297-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07297-4