Abstract
Helicobacter pylori, a member of the clade campylobacteria, is the leading cause of chronic gastritis and gastric cancer. Virulence and antibiotic resistance of H. pylori are of great concern to public health. However, the relationship between virulence and antibiotic resistance genes in H. pylori in relation to other campylobacteria remains unclear. Using the virulence and comprehensive antibiotic resistance databases, we explored all available 354 complete genomes of H. pylori and compared it with 90 species of campylobacteria for virulence and antibiotic resistance genes/proteins. On average, H. pylori had 129 virulence genes, highest among Helicobacter spp. and 71 antibiotic resistance genes, one of the lowest among campylobacteria. Just 2.6% of virulence genes were shared by all campylobacterial members, whereas 9.4% were unique to H. pylori. The cytotoxin-associated genes (cags) seemed to be exclusive to H. pylori. Majority of the isolates from Asia and South America were cag2-negative and many antibiotic resistance genes showed isolate-specific patterns of occurrence. Just 15 (8.8%) antibiotic resistance genes, but 103 (66%) virulence genes including 25 cags were proteomically identified in H. pylori. Arcobacterial members showed large variation in the number of antibiotic resistance genes and there was a positive relation with the genome size. Large repository of antibiotic resistance genes in campylobacteria and a unique set of virulence genes might have important implications in sha** the course of virulence and antibiotic resistance in H. pylori.



Similar content being viewed by others
Data Availability
The sequence data used in this work were obtained from NCBI. The relevant derived data are given in the supplemental result tables accessible at https://osf.io/rmd9y/.
References
Hooi JKY, Lai WY, Ng WK et al (2017) Global prevalence of Helicobacter pylori infection: Systematic review and meta-analysis. Gastroenterol 153:P420–P429. https://doi.org/10.1053/j.gastro.2017.04.022
Suerbaum S, Michetti P (2002) Helicobacter pylori infection. N Engl J Med 347:1175–1186. https://doi.org/10.1056/NEJMra020542
Etemadi A, Safiri S, Sepanlou SG et al (2020) The global, regional, and national burden of stomach cancer in 195 countries, 1990–2017: A systematic analysis for the Global Burden of Disease study 2017. Lancet Gastroenterol Hepatol 5:42–54. https://doi.org/10.1016/S2468-1253(19)30328-0
Chiang T-H, Chang W-J, Chen SL-S et al (2021) Mass eradication of Helicobacter pylori to reduce gastric cancer incidence and mortality: A long-term cohort study on Matsu Islands. Gut 70:243–250. https://doi.org/10.1136/gutjnl-2020-322200
Choi IJ, Kim CG, Lee JY et al (2020) Family history of gastric cancer and Helicobacter pylori treatment. N Engl J Med 382:427–436. https://doi.org/10.1056/NEJMoa1909666
Malfertheiner P, Megraud F, O’Morain CA et al (2017) Management of Helicobacter pylori infection—the Maastricht V/Florence Consensus Report. Gut 66:6–30. https://doi.org/10.1136/gutjnl-2016-312288
Pasceri V, Cammarota G, Patti G et al (1998) Association of virulent Helicobacter pylori strains with ischemic heart disease. Circulation 97:1675–1679. https://doi.org/10.1161/01.CIR.97.17.1675
Hu Y, Zhu Y, Lu N-H (2017) Novel and effective therapeutic regimens for Helicobacter pylori in an era of increasing antibiotic resistance. Front Cell Infect Microbiol 7:168. https://doi.org/10.3389/fcimb.2017.00168
Savoldi A, Carrara E, Graham DY et al (2018) Prevalence of antibiotic resistance in Helicobacter pylori: a systematic review and meta-analysis in World Health Organization regions. Gastroenterol 155:1372-1382.e17. https://doi.org/10.1053/j.gastro.2018.07.007
Tacconelli E, Carrara E, Savoldi A et al (2018) Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. https://doi.org/10.1016/S1473-3099(17)30753-3
Gingold-Belfer R, Niv Y, Schmilovitz-Weiss H et al (2021) Susceptibility-guided versus empirical treatment for Helicobacter pylori infection: a systematic review and meta-analysis. J Gastroenterol Hepatol 36:2649–2658. https://doi.org/10.1111/jgh.15575
Megraud F, Bruyndonckx R, Coenen S et al (2021) Helicobacter pylori resistance to antibiotics in Europe in 2018 and its relationship to antibiotic consumption in the community. Gut 70:1815–1822. https://doi.org/10.1136/gutjnl-2021-324032
Nestegard O, Moayeri B, Halvorsen F-A et al (2022) Helicobacter pylori resistance to antibiotics before and after treatment: Incidence of eradication failure. PLoS ONE 17:e0265322. https://doi.org/10.1371/journal.pone.0265322
Shu X, Ye D, Hu C et al (2022) Alarming antibiotics resistance of Helicobacter pylori from children in Southeast China over 6 years. Sci Rep 12:17754. https://doi.org/10.1038/s41598-022-21661-y
Brennan DE, Dowd C, O’Morain C et al (2018) Can bacterial virulence factors predict antibiotic resistant Helicobacter pylori infection? World J Gastroenterol 24:971–981. https://doi.org/10.3748/wjg.v24.i9.971
da Silva Benigno TG, Ribeiro Junior HL, de Azevedo OGR et al (2022) Clarithromycin-resistant H. pylori primary strains and virulence genotypes in the Northeastern region of Brazil. Revista do Instituto de Medicina Tropical de São Paulo 64:e47. https://doi.org/10.1590/s1678-9946202264047
Hosseini RS, Rahimian G, Shafigh MH et al (2021) Correlation between clarithromycin resistance, virulence factors and clinical characteristics of the disease in Helicobacter pylori infected patients in Shahrekord. Southwest Iran AMB Express 11:147. https://doi.org/10.1186/s13568-021-01310-9
Liu Y, Wang S, Yang F et al (2022) Antimicrobial resistance patterns and genetic elements associated with the antibiotic resistance of Helicobacter pylori strains from Shanghai. Gut Pathogens 14:14. https://doi.org/10.1186/s13099-022-00488-y
Igwaran A, Okoh AI (2019) Human campylobacteriosis: a public health concern of global importance. Heliyon 5:e02814. https://doi.org/10.1016/j.heliyon.2019.e02814
Kaakoush NO, Castaño-Rodríguez N, Mitchell HM, Man SM (2015) Global epidemiology of Campylobacter infection. Clin Microbiol Rev 28:687–720. https://doi.org/10.1128/CMR.00006-15
Ferreira EO, Lagacé-Wiens P, Klein J (2022) Campylobacter concisus gastritis masquerading as Helicobacter pylori on gastric biopsy. Helicobacter 27:e12864. https://doi.org/10.1111/hel.12864
Hlashwayo DF, Sigaúque B, Noormahomed EV et al (2021) A systematic review and meta-analysis reveal that Campylobacter spp. and antibiotic resistance are widespread in humans in sub-Saharan Africa. PLoS ONE 16:e0245951. https://doi.org/10.1371/journal.pone.0245951
EFSA (2022) The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J 20:7209. https://doi.org/10.2903/j.efsa.2022.7209
Wang D, Guo Q, Yuan Y, Gong Y (2019) The antibiotic resistance of Helicobacter pylori to five antibiotics and influencing factors in an area of China with a high risk of gastric cancer. BMC Microbiol 19:152. https://doi.org/10.1186/s12866-019-1517-4
Alcock BP, Raphenya AR, Lau TTY et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. https://doi.org/10.1093/nar/gkz935
Liu B, Zheng D, ** Q et al (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. https://doi.org/10.1093/nar/gky1080
Feldgarden M, Brover V, Gonzalez-Escalona N et al (2021) AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep 11:12728. https://doi.org/10.1038/s41598-021-91456-0
Liu B, Pop M (2009) ARDB—antibiotic resistance genes database. Nucleic Acids Res 37:D443–D447. https://doi.org/10.1093/nar/gkn656
Doster E, Lakin SM, Dean CJ et al (2020) MEGARes 2.0: a database for classification of antimicrobial drug, biocide and metal resistance determinants in metagenomic sequence data. Nucleic Acids Res 48:D561–D569. https://doi.org/10.1093/nar/gkz1010
Bortolaia V, Kaas RS, Ruppe E et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. https://doi.org/10.1093/jac/dkaa345
Hendriksen RS, Bortolaia V, Tate H et al (2019) Using genomics to track global antimicrobial resistance. Front Public Health 7:242. https://doi.org/10.3389/fpubh.2019.00242
Rao RSP, Ghate SD, Shastry RP et al (2023) Prevalence and heterogeneity of antibiotic resistance genes in Orientia tsutsugamushi and other rickettsial genomes. Microb Pathog 174:105953. https://doi.org/10.1016/j.micpath.2022.105953
Zhang Z, Zhang Q, Wang T et al (2022) Assessment of global health risk of antibiotic resistance genes. Nat Commun 13:1553
Mootapally C, Mahajan MS, Nathani NM (2021) Sediment plasmidome of the Gulfs of Kathiawar Peninsula and Arabian Sea: insights gained from metagenomics data. Microb Ecol 81:540–548. https://doi.org/10.1007/s00248-020-01587-6
Tamura K, Stecher G, Kumar S (2021) MEGA 11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Agresti A (2018) An introduction to categorical data analysis, 3rd edn. Wiley, New York
Vijaymeena MK, Kavitha K (2016) A survey on similarity measures in text mining. Mach Learn Appl 3:19–28. https://doi.org/10.5121/mlaij.2016.3103
Karlsson R, Thorell K, Hosseini S et al (2016) Comparative analysis of two Helicobacter pylori strains using genomics and mass spectrometry-based proteomics. Front Microbiol 7:1757. https://doi.org/10.3389/fmicb.2016.01757
Kumar S, Schmitt C, Gorgette O et al (2022) Bacterial membrane vesicles as a novel strategy for extrusion of antimicrobial bismuth drug in Helicobacter pylori. MBio 13:e01633-e1722. https://doi.org/10.1128/mbio.01633-22
Loh JT, Shum MV, Jossart SD et al (2021) Delineation of the pH-responsive regulon controlled by the Helicobacter pylori ArsRS two-component system. Infect Immun 89:e00597-e620. https://doi.org/10.1128/IAI.00597-20
Müller SA, Findeiß S, Pernitzsch SR et al (2013) Identification of new protein coding sequences and signal peptidase cleavage sites of Helicobacter pylori strain 26695 by proteogenomics. J Proteom 86:27–42. https://doi.org/10.1016/j.jprot.2013.04.036
Müller SA, Pernitzsch SR, Haange SB et al (2015) Stable isotope labeling by amino acids in cell culture based proteomics reveals differences in protein abundances between spiral and coccoid forms of the gastric pathogen Helicobacter pylori. J Proteom 126:34–45. https://doi.org/10.1016/j.jprot.2015.05.011
Sugiyama N, Miyake S, Lin MH et al (2019) Comparative proteomics of Helicobacter pylori strains reveals geographical features rather than genomic variations. Genes Cells 24:139–150. https://doi.org/10.1111/gtc.12662
Wei S, Li S, Wang J et al (2022) Outer membrane vesicles secreted by Helicobacter pylori transmitting gastric pathogenic virulence factors. ACS Omega 7:240–258. https://doi.org/10.1021/acsomega.1c04549
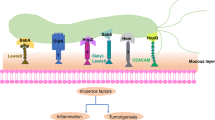
Censini S, Lange C, **ang Z et al (1996) Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA 93:14648–14653. https://doi.org/10.1073/pnas.93.25.14648
Collado L, Figueras MJ (2011) Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin Microbiol Rev 24:174–192. https://doi.org/10.1128/CMR.00034-10
Pérez-Cataluña A, Salas-Massó N, Diéguez AL et al (2018) Revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front Microbiol 9:2077. https://doi.org/10.3389/fmicb.2018.02077
Larsson DGJ, Flach C-F (2022) Antibiotic resistance in the environment. Nat Rev Microbiol 20:257–269. https://doi.org/10.1038/s41579-021-00649-x
Zhang Q, Zhang Z, Lu T et al (2020) Cyanobacterial blooms contribute to the diversity of antibiotic-resistance genes in aquatic ecosystems. Commun Biol 3:737. https://doi.org/10.1038/s42003-020-01468-1
Noto JM, Peek RM Jr (2012) The Helicobacter pylori cag pathogenicity island. Methods Mol Biol 921:41–50. https://doi.org/10.1007/978-1-62703-005-2_7
Hatakeyama M (2017) Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc Jpn Acad B Phys Biol Sci 93:196–219. https://doi.org/10.2183/pjab.93.013
Yu M, Xu M, Shen Y et al (2023) Hp0521 inhibited the virulence of H. pylori 26,695 strain via regulating CagA expression. Heliyon 9:e17881. https://doi.org/10.1016/j.heliyon.2023.e17881
Yuan X-y, Yan J-J, Yang Y-c et al (2017) Helicobacter pylori with East Asian-type cagPAI genes is more virulent than strains with Western-type in some cagPAI genes. Braz J Microbiol 48:218–224. https://doi.org/10.1016/j.bjm.2016.12.004
Geisinger E, Isberg RR (2017) Interplay between antibiotic resistance and virulence during disease promoted by multidrug-resistant bacteria. J Infect Dis 215:S9–S17. https://doi.org/10.1093/infdis/jiw402
Abdi SN, Ghotaslou R, Ganbarov K et al (2020) Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect Drug Resist 13:423–434. https://doi.org/10.2147/IDR.S228089
Nikaido H (2009) Multidrug resistance in bacteria. Annu Rev Biochem 78:119–146. https://doi.org/10.1146/annurev.biochem.78.082907.145923
Salini S, Muralikrishnan B, Bhat SG et al (2022) Overexpression of a membrane transport system MSMEG_1381 and MSMEG_1382 confers multidrug resistance in Mycobacterium smegmatis. Preprints 202204.0003.v2. https://doi.org/10.20944/preprints202204.0003.v2.
van Hoek AHAM, Mevius D, Guerra B et al (2011) Acquired antibiotic resistance genes: an overview. Front Microbiol 2:203. https://doi.org/10.3389/fmicb.2011.00203
Kent AG, Vill AC, Shi Q et al (2020) Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi–C. Nat Commun 11:4379. https://doi.org/10.1038/s41467-020-18164-7
Oyarzabal OA, Rad R, Backert S (2007) Conjugative transfer of chromosomally encoded antibiotic resistance from Helicobacter pylori to Campylobacter jejuni. J Clin Microbiol 45:402–408. https://doi.org/10.1128/JCM.01456-06
Stingl K, Müller S, Scheidgen-Kleyboldt G et al (2010) Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc Natl Acad Sci USA 107:1184–1189. https://doi.org/10.1073/pnas.0909955107
Her H-L, Lin P-T, Wu Y-W (2021) PangenomeNet: A pan-genome-based network reveals functional modules on antimicrobial resistome for Escherichia coli strains. BMC Bioinformatics 22:548. https://doi.org/10.1186/s12859-021-04459-z
Acknowledgements
NA gratefully acknowledges the initial funding support from the OU VPRP office for the establishment of the Proteomics Core Facility.
Funding
This work did not receive any specific funding.
Author information
Authors and Affiliations
Contributions
RSPR and SDG planned and performed the work, and wrote the manuscript. LP helped in data curation. All authors contributed intellectually, and edited/reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical Approval
The work is in compliance with ethical standards. No ethical clearance was necessary.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rao, R.S.P., Ghate, S.D., Pinto, L. et al. Extent of Virulence and Antibiotic Resistance Genes in Helicobacter pylori and Campylobacteria. Curr Microbiol 81, 154 (2024). https://doi.org/10.1007/s00284-024-03653-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-024-03653-5