Abstract
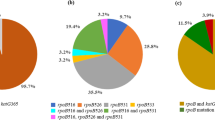
Drug resistance to streptomycin in the clinical isolates of Mycobacterium tuberculosis (MTB) needs special consideration. It can mostly be caused by mutations in four genes with the names rpsL, rrs, gidB, and whiB7. The main objective of this study was the evaluation of the type and frequency of mutations in these mentioned genes using the PCR-sequencing method. This study was performed on 15 streptomycin-resistant and five streptomycin-sensitive isolates. Among resistant isolates, 11 samples contained mutations in codon 43 of the rpsL gene, which caused the lysine to be converted to arginine. Additionally, all of the isolates had mutations in the gidB. Missense mutations in codons 92 and 20 of this gene result in the amino acids Glutamic acid or Arginine being changed to Aspartic acid or Proline, respectively. No mutations in the rrs or whiB7 were found in any of the samples. Simultaneous mutations of rpsL and gidB were found in 10 isolates, the majority of which were Bei**g strain. The results showed that the mutations of rpsL and gidB genes are mostly responsible for the streptomycin resistance in the evaluated MTB isolates. Furthermore, the discovery of dual mutations in Bei**g strains highlights the strain's considerable potential for develo** Tuberculosis drug resistance.

Similar content being viewed by others
Data Availability
Data transparency is provided.
Code Availability
Not applicable.
References
Gagneux S (2018) Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 16(4):202
Reid MJ, Arinaminpathy N, Bloom A, Bloom BR, Boehme C, Chaisson R, Chin DP, Churchyard G, Cox H, Ditiu L (2019) Building a tuberculosis-free world: the lancet commission on tuberculosis. Lancet 393(10178):1331–1384
World Health Organization (WHO) (2020) Global Tuberculosis Report 2020. https://www.who.int/publications/i/item/9789240013131
Paleckyte A, Dissanayake O, Mpagama S, Lipman MC, McHugh TD (2021) Reducing the risk of tuberculosis transmission for HCWs in high incidence settings. Antimicrob Resist Infect Control 10(1):1–11
Moore DA, Evans CA, Gilman RH, Caviedes L, Coronel J, Vivar A, Sanchez E, Piñedo Y, Saravia JC, Salazar C (2006) Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med 355(15):1539–1550
Miotto P, Zhang Y, Cirillo DM, Yam WC (2018) Drug resistance mechanisms and drug susceptibility testing for tuberculosis. Respirology 23(12):1098–1113
Gaude GS, Hattiholli J, Kumar P (2014) Risk factors and drug-resistance patterns among pulmonary tuberculosis patients in northern Karnataka region, India. Niger Med J 55(4):327
World Health Organization (WHO) (2008) WHO/IUATLD global project on anti-tuberculosis drug resistance surveillance. https://apps.who.int/iris/bitstream/handle/10665/43831/9789241563543_eng.pdf?sequence=1
Kerantzas CA, Jacobs WR (2017) Origins of combination therapy for tuberculosis: lessons for future antimicrobial development and application. MBio. https://doi.org/10.1128/mBio.01586-16
Karimi S, Mirhendi H, Zaniani FR, Manesh SE, Salehi M, Esfahani BN (2017) Rapid Detection of streptomycin-Resistant Mycobacterium tuberculosis by rpsL-restriction fragment length polymorphism. Adv biomed res. https://doi.org/10.4103/abr.abr_240_16
Jagielski T, Ignatowska H, Bakuła Z, Dziewit Ł, Napiórkowska A, Augustynowicz-Kopeć E, Zwolska Z, Bielecki J (2014) Screening for streptomycin resistance-conferring mutations in Mycobacterium tuberculosis clinical isolates from Poland. PLoS ONE 9(6):e100078
Okamoto S, Tamaru A, Nakajima C, Nishimura K, Tanaka Y, Tokuyama S, Suzuki Y, Ochi K (2007) Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol Microbiol 63(4):1096–1106
Bwalya P, Yamaguchi T, Solo ES, Chizimu JY, Mbulo G, Nakajima C, Suzuki Y (2021) Characterization of mutations associated with streptomycin resistance in multidrug-resistant Mycobacterium tuberculosis in Zambia. Antibiotics 10(10):1169
Wang Y, Li Q, Gao H, Zhang Z, Liu Y, Lu J, Dai E (2019) The roles of rpsL, rrs, and gidB mutations in predicting streptomycin-resistant drugs used on clinical Mycobacterium tuberculosis isolates from Hebei Province. China Int J Clin Exp Pathol 12(7):2713
Tudó G, Rey E, Borrell S, Alcaide F, Codina G, Coll P, Martín-Casabona N, Montemayor M, Moure R, Orcau A (2010) Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis clinical isolates in the area of Barcelona. J Antimicrob Chemother 65(11):2341–2346
Reeves AZ, Campbell PJ, Sultana R, Malik S, Murray M, Plikaytis BB, Shinnick TM, Posey JE (2013) Aminoglycoside cross-resistance in Mycobacterium tuberculosis due to mutations in the 5′ untranslated region of whiB7. Antimicrob Agents Chemother 57(4):1857–1865
Villellas C, Aristimuño L, Vitoria M-A, Prat C, Blanco S, de Viedma DG, Domínguez J, Samper S, Aínsa JA (2013) Analysis of mutations in streptomycin-resistant strains reveals a simple and reliable genetic marker for identification of the Mycobacterium tuberculosis Bei**g genotype. J Clin Microbiol 51(7):2124–2130
Shafipour M, Shirzad-Aski H, Ghaemi EA, Sohrabi A, Babaii Kochaksaraei M, Taziki M, Rahimi S, Ghazvini K, Baei B (2021) Mycobacterium tuberculosis ty** using allele-specific oligonucleotide multiplex PCR (ASO–PCR) method. Curr Microbiol 8(12):4009–4013
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, De Haas P, Van Deutekom H, Roring S (2006) Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat ty** of Mycobacterium tuberculosis. J Clin Microbiol 44(12):4498–4510
World Health Organization (WHO) Dose optimization of rifampicin, isoniazid, pyrazinamide and ethambutol in the treatment of drug-susceptible tuberculosis. https://www.who.int/docs/default-source/hq-tuberculosis/dose-optimization-concept-note.pdf?sfvrsn=934ab3e2_2
The Centers for Disease Control and Prevention (CDC) (2022) Surveillance definitions for extensively drug resistant (XDR) and pre-XDR tuberculosis. CDC. https://www.cdc.gov/tb/publications/letters/2022/surv-def-xdr.html
Hlaing YM, Tongtawe P, Tapchaisri P, Thanongsaksrikul J, Thawornwan U, Archanachan B, Srimanote P (2017) Mutations in streptomycin resistance genes and their relationship to streptomycin resistance and lineage of Mycobacterium tuberculosis Thai isolates. Tuberc Respir Dis 80(2):159
World Health Organization (2019) WHO consolidated guidelines on drug-resistant tuberculosis treatment (No. WHO/CDS/TB/2019.7). World Health Organization
Cohen KA, Stott KE, Munsamy V, Manson AL, Earl AM, Pym AS (2020) Evidence for expanding the role of streptomycin in the management of drug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother 64(9):e00860-e820
Shrestha D, Maharjan B, Oo NAT, Isoda N, Nakajima C, Suzuki Y (2020) Molecular analysis of streptomycin-resistance associating genes in Mycobacterium tuberculosis isolates from Nepal. Tuberculosis 125:101985
Khosravi AD, Etemad N, Hashemzadeh M, Dezfuli SK, Goodarzi H (2017) Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J Glob Antimicrob Resist 9:51–56
Nhu N, Lan N, Phuong N, van Chau NV, Farrar J, Caws M (2012) Association of streptomycin resistance mutations with level of drug resistance and Mycobacterium tuberculosis genotypes. Int J Tuberc Lung Dis 16(4):527–531
Zhao L-l, Liu H-c, Sun Q, **ao T-y, Zhao X-q, Li G-l, Zeng C-y, Wan K-l (2015) Identification of mutations conferring streptomycin resistance in multidrug-resistant tuberculosis of China. Diagn Microbiol Infect Dis 83(2):150–153
Sun Y-J, Luo J-T, Wong S-Y, Lee A (2010) Analysis of rpsL and rrs mutations in Bei**g and non-Bei**g streptomycin-resistant Mycobacterium tuberculosis isolates from Singapore. Clin Microbiol Infect 16(3):287–289
Smittipat N, Juthayothin T, Billamas P, Jaitrong S, Rukseree K, Dokladda K, Chaiyasirinroje B, Disratthakit A, Chaiprasert A, Mahasirimongkol S (2016) Mutations in rrs, rpsL and gidB in streptomycin-resistant Mycobacterium tuberculosis isolates from Thailand. J Glob Antimicrob Resist 4:5–10
Sun H, Zhang C, **ang L, Pi R, Guo Z, Zheng C, Li S, Zhao Y, Tang K, Luo M (2016) Characterization of mutations in streptomycin-resistant Mycobacterium tuberculosis isolates in Sichuan, China and the association between Bei**g-lineage and dual-mutation in gidB. Tuberculosis 96:102–106
Acknowledgements
We thank the staff of the Department of Microbiology, which belonged to Golestan University of Medical Sciences, for their kind cooperation.
Funding
The work was supported by Golestan University of Medical Sciences, Gorgan, Iran (grant number: IR.GOUMS.111209).
Author information
Authors and Affiliations
Contributions
(i) The conception or design of the study: EAG, HS, MS. (ii) the acquisition: MS, HS, SZ, KG analysis: HS, MS interpretation of the data: HS, MS, SG, AM, PMK; and (iii) writing of the manuscript: HS, MS, EAG. All authors approved the final version.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare for this study.
Ethics Approval
This study was approved by the Ethical Committee of Golestan University of Medical Sciences, Iran (Ethical code: IR.GOUMS.REC.1399.331).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shafipour, M., Shirzad-Aski, H., Mohammadzadeh, A. et al. Evaluation of Mutations Related to Streptomycin Resistance in Mycobacterium tuberculosis Clinical Isolates. Curr Microbiol 79, 343 (2022). https://doi.org/10.1007/s00284-022-03043-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-03043-9