Abstract
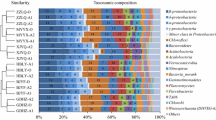
An active microbial community of nitrifying and denitrifying bacteria is needed for efficient utilization of nitrogenous compounds from wastewater. In this study, we explored the bacterial community diversity and structure within rivers, treated and untreated wastewater treatment plants (WWTPs) discharging into Lake Victoria. Water samples were collected from rivers and WWTPs that drain into Lake Victoria. Physicochemical analysis was done to determine the level of nutrients or pollutant loading in the samples. Total community DNA was extracted, followed by Illumina high throughput sequencing to determine the total microbial community and abundance. Enrichment and isolation were then done to recover potential nitrifiers and denitrifiers. Physicochemical analysis pointed to high levels total nitrogen and ammonia in both treated and untreated WWTPs as compared to the samples from the lake and rivers. A total of 1,763 operational taxonomic units (OTUs) spread across 26 bacterial phyla were observed with the most dominant phylum being Proteobacteria. We observed a decreasing trend in diversity from the lake, rivers to WWTPs. The genus Planktothrix constituted 19% of the sequence reads in sample J2 collected from the lagoon. All the isolates recovered in this study were affiliated to three genera: Pseudomonas, Klebsiella and Enterobacter in the phylum Proteobacteria. A combination of metagenomic analysis and a culture-dependent approach helped us understand the relative abundance as well as potential nitrifiers and denitrifiers present in different samples. The recovered isolates could be used for in situ removal of nitrogenous compounds from contaminated wastewater.





Similar content being viewed by others
Data Availability
The Raw sequence reads have been deposited into the SRA under the accession PRJNA824944. Sequence reads from the Illumina sequencing are under the Biosample SUB170771 while the isolate sequences are under the Biosample SUB11316194 (Accession numbers ON227417-ON227434).
References
Galloway JN, Winiwarter W, Leip A et al (2014) Nitrogen footprints: past, present and future. Environ Res Lett. https://doi.org/10.1088/1748-9326/9/11/115003
Gao Y, Zhou F, Ciais P et al (2020) Human activities aggravate nitrogen-deposition pollution to inland water over China. Natl Sci Rev. https://doi.org/10.1093/nsr/nwz073
Sutton M, Mason K, Bleeker A et al (2020) Just Enough Nitrogen: Perspectives on how to get there for regions with too much and too little nitrogen. Springer International Publishing, Cham
Ghafari S, Hasan M, Aroua MK (2008) Bio-electrochemical removal of nitrate from water and wastewater-a review. Bioresou Technol. https://doi.org/10.1016/j.biortech.2007.05.026
le Moal M, Gascuel-Odoux C, Ménesguen A et al (2019) Eutrophication: A new wine in an old bottle? Sci Total Environ. https://doi.org/10.1016/j.scitotenv.2018.09.139
Juma DW, Wang H, Li F (2014) Impacts of population growth and economic development on water quality of a lake: case study of Lake Victoria Kenya water. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-014-2524-5
Opande GO, Onyango JC, Wagai SO (2004) Lake Victoria: The water hyacinth (Eichhornia crassipes [MART.] SOLMS), its socio-economic effects, control measures and resurgence in the Winam gulf. Limnologica. https://doi.org/10.1016/S0075-9511(04)80028-8
Wanda FM, Namukose M, Matuha M (2015) Water hyacinth hotspots in the Ugandan waters of Lake Victoria in 1994–2012: implications for management. Afr J Aquat Sci. https://doi.org/10.2989/16085914.2014.997181
Mbonde AS, Sitoki L, Kurmayer R (2015) Phytoplankton composition and microcystin concentrations in open and closed bays of Lake Victoria Tanzania. Aquat Ecosyst Health Manag. https://doi.org/10.1080/14634988.2015.1011030
Shukla S, Saxena A (2019) Global status of nitrate contamination in groundwater: its occurrence, health impacts, and mitigation measures. In: Hussain CM (ed) Handbook of environmental materials management. Springer International Publishing, Chem
Khardenavis AA, Kapley A, Purohit HJ (2007) Simultaneous nitrification and denitrification by diverse Diaphorobacter sp. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-007-1176-5
Zhang Y, Song C, Zhou Z et al (2019) Coupling between nitrification and denitrification as well as its effect on phosphorus release in sediments of Chinese shallow lakes. Water (Switzerland). https://doi.org/10.3390/w11091809
Cho S, Kambey C, Nguyen VK (2020) Performance of anammox processes for wastewater treatment: a critical review on effects of operational conditions and environmental stresses. Water (Switzerland) 12(1):20
Do TT, Delaney S, Walsh F (2019) 16S rRNA gene based bacterial community structure of wastewater treatment plant effluents. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnz017
Ji B, Yang K, Zhu L et al (2015) Aerobic denitrification: A review of important advances of the last 30 years. Biotechnol Bioprocess Eng. https://doi.org/10.1007/s12257-015-0009-0
Lei X, Jia Y, Chen Y, Hu Y (2019) Simultaneous nitrification and denitrification without nitrite accumulation by a novel isolated Ochrobactrum anthropic LJ81. Biores Technol. https://doi.org/10.1016/j.biortech.2018.10.060
Joo HS, Hirai M, Shoda M (2005) Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis no. 4. J Biosci Bioeng. https://doi.org/10.1263/jbb.100.184
Ruan Y, Taherzadeh MJ, Kong D et al (2020) Nitrogen removal performance and metabolic pathways analysis of a novel aerobic denitrifying halotolerant pseudomonas balearica strain RAD-17. Microorganisms. https://doi.org/10.3390/microorganisms8010072
APHA (2005) Standard methods for the examination of water and wastewater. Standard Methods, Washington
Kiplimo D, Mugweru J, Kituyi S et al (2019) Diversity of esterase and lipase producing haloalkaliphilic bacteria from Lake Magadi in Kenya. J Basic Microbiol. https://doi.org/10.1002/jobm.201900353
Caporaso JG, Lauber CL, Walters WA et al (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1000080107
Edgar RC, Haas BJ, Clemente JC et al (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. https://doi.org/10.1093/bioinformatics/btr381
Ssekagiri AT, Sloan W, Zeeshan Ijaz U (2017) microbiomeSeq: an R package for analysis of microbial communities in an environmental context. ISCB Africa ASBCB conference, Entebbe
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. https://doi.org/10.1371/journal.pone.0061217
Oksanen J, Blanchet FG, Friendly M et al (2015) Vegan community ecology package Ordination methods, diversity analysis and other functions for community and vegetation ecologists. R package version 2:2
Warnes GR, Bolker B, Bonebakker L, et al (2016) Package “gplots”: Various R programming tools for plotting data. R package version 2170
Andersen K, Kirkegaard R, Karst S et al (2018) ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. Org BioRxiv. https://doi.org/10.1101/299537
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. https://doi.org/10.1093/molbev/msy096
Jewell TNM, Karaoz U, Bill M et al (2017) Metatranscriptomic analysis reveals unexpectedly diverse microbial metabolism in a biogeochemical hot spot in an alluvial aquifer. Front Microbiol. https://doi.org/10.3389/fmicb.2017.00040
Yi XH, Wan J, Ma Y et al (2017) Structure and succession of bacterial communities of the granular sludge during the initial stage of the simultaneous denitrification and methanogenesis Process. Water Air Soil Pollut. https://doi.org/10.1007/s11270-016-3168-5
Wexler M, Bond PL, Richardson DJ, Johnston AWB (2011) Functional screening a wide host-range metagenomic library from a wastewater treatment plant yields a novel alcohol/aldehyde dehydrogenase. In: de Bruijn FJ (ed) Handbook of molecular microbial ecology II: metagenomics in different habitats. John Wiley, Hoboken
Marie B, Huet H, Marie A et al (2012) Effects of a toxic cyanobacterial bloom (Planktothrix agardhii) on fish: insights from histopathological and quantitative proteomic assessments following the oral exposure of medaka fish (Oryzias latipes). Aquat Toxicol. https://doi.org/10.1016/j.aquatox.2012.02.008
Legendre P, de Cáceres M (2013) Beta diversity as the variance of community data: dissimilarity coefficients and partitioning. Ecol Lett. https://doi.org/10.1111/ele.12141
Chakraborty A, Bhadury P (2015) Effect of pollution on aquatic microbial diversity. In: Sukla LB, Pradhan N, Panda S, Mishra BK (eds) Environmental microbial biotechnology. Springer International Publishing, Chem
Xu J, Wang P, Li Y et al (2019) Shifts in the microbial community of activated sludge with different COD/N ratios or dissolved oxygen levels in Tibet. China Sustainability (Switzerland). https://doi.org/10.3390/su11082284
Soltani N, Khodaei K, Alnajar N et al (2012) Cyanobacterial community patterns as water quality bioindicators. Iran J Fish Sci 11(4):876–891
Nascimento AL, Souza AJ, Andrade PAM et al (2018) Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front Microbiol. https://doi.org/10.3389/fmicb.2018.01462
Newton RJ, Mcmahon KD (2011) Seasonal differences in bacterial community composition following nutrient additions in a eutrophic lake. Environ Microbiol. https://doi.org/10.1111/j.1462-2920.2010.02387.x
Lu H, Chandran K, Stensel D (2014) Microbial ecology of denitrification in biological wastewater treatment. Water Res. https://doi.org/10.1016/j.watres.2014.06.042
Ge S, Wang S, Yang X et al (2015) Detection of nitrifiers and evaluation of partial nitrification for wastewater treatment: a review. Chemosphere. https://doi.org/10.1016/j.chemosphere.2015.02.004
Sun D, Tang X, Zhao M et al (2020) Distribution and diversity of comammox nitrospira in coastal wetlands of China. Front Microbiol. https://doi.org/10.3389/fmicb.2020.589268
Wan W, He D, Xue Z (2017) Removal of nitrogen and phosphorus by heterotrophic nitrification-aerobic denitrification of a denitrifying phosphorus-accumulating bacterium enterobacter cloacae HW-15. Ecol Eng. https://doi.org/10.1016/j.ecoleng.2016.11.030
Feng Y, Feng J, Shu QL (2018) Isolation and characterization of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae and Klebsiella variicola strains from various environments. J Appl Microbiol. https://doi.org/10.1111/jam.13703
Motamedi H, Jafari M (2020) Screening heterotrophic ammonia removal and aerobic denitrifying bacteria from wastewater of ammonia production units of a petrochemical industry. Curr Microbiol. https://doi.org/10.1007/s00284-020-02065-5
Su W, Zhang L, Li D et al (2012) Dissimilatory nitrate reduction by Pseudomonas alcaliphila with an electrode as the sole electron donor. Biotechnol Bioeng. https://doi.org/10.1002/bit.24554
Zhang W, Zhang Y, Su W et al (2014) Effects of cathode potentials and nitrate concentrations on dissimilatory nitrate reductions by Pseudomonas alcaliphila in bioelectrochemical systems. J Environ Sci (China). https://doi.org/10.1016/S1001-0742(13)60460-X
Acknowledgements
James Murimi Wachira is grateful to the International Institute of Tropical Agriculture (IITA) and the University of Embu (UoEM) for their great support and contribution towards the entire research project.
Funding
Graduate Research Fellowship by the International Institute of Tropical Agriculture, National Research Fund, Kenya, [Grant number NRF/1/MMC/470,2018] and The Alexander von Humboldt Equipment Grant [Grant number Ref 3.4-8151/19060].
Author information
Authors and Affiliations
Contributions
JMW did the experiments, data collection and wrote the manuscript. DM assisted in data collection. MT and CM designed the experiments, guided sample collection, and proofread the manuscript. RM-guided data analysis, manuscript writing and proofreading. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Consent for Publication
The authors give consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2022_2950_MOESM1_ESM.png
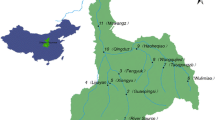
Supplementary file1 (PNG 294 kb)—Map showing the study area and sample collection sites in the vicinity of Lake Victoria
Rights and permissions
About this article
Cite this article
Wachira, J.M., Kiplimo, D., Thuita, M. et al. Community Structure of Nitrifying and Denitrifying Bacteria from Effluents Discharged into Lake Victoria, Kenya. Curr Microbiol 79, 252 (2022). https://doi.org/10.1007/s00284-022-02950-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02950-1