Abstract
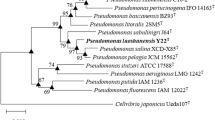
Strain 16W4-4-3 T was isolated from the oil-well production water in Qinghai Oilfield, China. Cells were Gram-stain-negative, rod-shaped, catalase- and oxidase-positive, facultatively anaerobic and motile by single polar flagellum. The 16S rRNA gene sequences of strain 16W4-4-3 T showed the highest similarities with Pseudomonas profundi M5T (98.8%), P. pelagia CL-AP6T (98.0%), P. salina XCD-X85T (97.7%), and P. sabulinigri J64T (97.5%). The phylogenetic trees based on multilocus sequence analyses with concatenating 16S rRNA, gyrB, rpoD and rpoB genes suggested that this strain should be affiliated to the genus Pseudomonas but remotely related from other species. In addition, whole genome analyses revealed that the digital DNA–DNA hybridization values and average nucleotide identities of strain 16W4-4-3 T against its close relatives were all below 28.8% and 86.5%, respectively. Furthermore, the isolate had totally different whole cell protein profile as compared to those of other species. Major fatty acids were summed feature 8 (C18:1ω7c and/or C18:1ω6c), C16:0, summed feature 3 (C16:1ω7c and/or C16:1ω6c) and C17:0cyclo. Major isoprenoid quinone was ubiquinone (Q-9), and major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, and phosphatidylethanolamine. The DNA G + C content was 58.5 mol%. Therefore, phenotypic, phylogenetic, genomic, chemotaxonomic, and proteomic traits showed that the isolate represented a novel species of the genus Pseudomonas, the name Pseudomonas saliphila sp. nov. is proposed. Type strain is 16W4-4-3 T (= CGMCC 1.13350 T = KCTC 72619 T).

Similar content being viewed by others
Abbreviations
- ANI:
-
Average nucleotide identity
- ANIb:
-
Average nucleotide identity with the BLASTN algorithm
- ANIm:
-
Average nucleotide identity using the MUMmer ultrarapid aligning tool
- dDDH:
-
Digital DNA–DNA hybridization
- MLSA:
-
Multilocus sequence analyses
- gyrB :
-
DNA gyrase, subunit B gene
- rpoB :
-
RNA polymerase beta subunit gene
- rpoD :
-
RNA polymerase sigma 70 (sigma D) factor gene
- MALDI-TOF MS:
-
Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- MA:
-
Marine agar 2216 (Difco)
- MB:
-
Marine broth 2216 (Difco
References
Palleroni NJ (2010) The pseudomonas story. Environ Microbiol 12:1377–1383. https://doi.org/10.1111/j.1462-2920.2009.02041.x
Lin SY, Hameed A, Liu YC, Hsu YH, Lai WA, Chen WM, Shen FT, Young CC (2013) Pseudomonas sagittaria sp. Nov., a siderophore-producing bacterium isolated from oil-contaminated soil. Int J Syst Evol Microbiol 63:2410. https://doi.org/10.1099/ijs.0.045567-0
Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E (2015) Phylogenomics and systematics in Pseudomonas. Front Microbiol 6:214. https://doi.org/10.3389/fmicb.2015.00214
Parte AC (2018) LPSN—list of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829. https://doi.org/10.1099/ijsem.0.002786
Mulet M, Gomila M, Lemaitre B, Lalucat J, García-Valdés E (2012) Taxonomic characterization of Pseudomonas strain L48 and formal proposal of Pseudomonas entomophila sp. nov. Syst Appl Microbiol 35:145–149. https://doi.org/10.1016/j.syapm.2011.12.003
Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E (2012) Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analyses approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol 35:455–464. https://doi.org/10.1016/j.syapm.2012.08.007
Peix A, Ramírez-Bahena MH, Velázquez E (2018) The current status on the taxonomy of Pseudomonas revisited: An update. Infect Genet Evol 57:106–116. https://doi.org/10.1016/j.meegid.2017.10.026
Tohya M, Watanabe S, Teramoto K, Shimojima M, Tada T, Kuwahara-Arai K, War MW, Mya S et al (2019) Pseudomonas juntendi sp. nov., isolated from patients in Japan and Myanmar. Int J Syst Evol Microbiol 69:3377–3384. https://doi.org/10.1099/ijsem.0.003623
Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. https://doi.org/10.1111/j.1574-6976.2011.00269.x
Cai M, Nie Y, Chi CQ, Tang YQ, Li Y, Wang XB, Liu ZS, Yang YF et al (2015) Crude oil as a microbial seed bank with unexpected functional potentials. Sci Rep 5:16057. https://doi.org/10.1038/srep16057
Tang SK, Li WJ, Wang D, Zhang YG, Xu LH et al (2003) Studies of the biological characteristics of some halophilic and halotolerant actinomycetes isolated from saline and alkaline soils. Actinomycetologica 17:6–10. https://doi.org/10.3209/saj.17_6
Tindall BJ, Sikorski J, Smibert RA, Krieg NR (2007) Phenotypic characterization and the principles of comparative systematics. In methods for general and molecular bacteriology, pp 341–352. Edited by Gerhardt P., et al. Washington, DC: American Society for Microbiology. doi: 10.1128/9781555817497.ch15
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluorescein. J Lab Clin Med 44:301–307
Meng Y, Liu H, Zhou Y, Cai M, Kang Y (2018) Vibrio gangliei sp. nov., a novel member of Vibrionaceae isolated from sawdust in a pigpen. Int J Syst Evol Microbiol 68:1969–1974. https://doi.org/10.1099/ijsem.0.002779
Menéndez E, Ramírez-Bahena MH, Fabryová A, Igual JM, Benada O, Mateos PF, Peix A, Kolařík M et al (2015) Pseudomonas coleopterorum sp. nov., a cellulase-producing bacterium isolated from the bark beetle Hylesinus fraxini. Int J Syst Evol Microbiol 65:2852–2858. https://doi.org/10.1099/ijs.0.000344
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Systematic Biol 20:406–416. https://doi.org/10.1093/sysbio/20.4.406
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analyses version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformat 14:60. https://doi.org/10.1186/1471-2105-14-60
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131. https://doi.org/10.1073/pnas.0906412106
Kew S, Banerjee T, Minihane AM, Finnegan YE, Williams CM, Calderet PC (2003) Relation between the fatty acid composition of peripheral blood mononuclear cells and measures of immune cell function in healthy, free-living subjects aged 25–72 y. Am J Clin Nutr 77:1278–1286. https://doi.org/10.1093/ajcn/77.5.1278
Komagata K, Suzuki KI (1987) Lipid and cell-wall analyses in bacterial systematics. Methods Microbiol 19:161–207. https://doi.org/10.1016/S0580-9517(08)70410-0
Kates M (1972) Techniques of lipidology: isolation, analysis and identification of lipids. Lab Tech Biochem Mol Biol 350:436–438. https://doi.org/10.1016/s0075-7535(08)70544-8
Shu L, Yang Y (2017) Bacillus classification based on matrix-assisted laser desorption ionization time-of-flight mass spectrometry—effects of culture conditions. Sci Rep 7:15546. https://doi.org/10.1038/s41598-017-15808-5
Hwang C, Zhang G, Kang S, Kim HJ, Cho BC (2009) Pseudomonas pelagia sp. nov., isolated from a culture of the Antarctic green alga Pyramimonas gelidicola. Int J Syst Evol Microbiol 59:3019–3024. https://doi.org/10.1099/ijs.0.008102-0
Kim K, Roh SW, Chang H, Nam Y, Yoon J, Jeon CO, Oh H, Bae J et al (2009) Pseudomonas sabulinigri sp. nov., isolated from black beach sand. Int J Syst Evol Microbiol 59:38–41. https://doi.org/10.1099/ijs.0.65866-0
Sun J, Wang W, Ying Y, Zhu X, Liu J (2018) Pseudomonas profundi sp. nov., isolated from deep-sea water. Int J Syst Evol Microbiol 68:1776–1780. https://doi.org/10.1099/ijsem.0.002748
Zhong Z, Liu Y, Hou T, Liu H, Zhou Y (2015) Pseudomonas salina sp. nov., isolated from a salt lake in Qaidam basin. China Int J Syst Evol Microbiol 65:2846–2851. https://doi.org/10.1099/ijs.0.000341
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MSD, Rooney AP, Yi H et al (2018) Proposed minimal standards for the use of the genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Acknowledgements
The authors thank the Institute of Microbiology, Chinese Academy of Sciences, for providing the transmission electron microscopy facility. The GenBank/EMBL/DDBJ accession numbers for whole genome and 16S rRNA gene sequences of strain 16W4-4-3 T are VTVA00000000 and MN423266; and whole genome accession numbers for Pseudomonas profundi M5T and P. salina CGMCC 1.12482 T are VTPZ00000000 and VTTI00000000, respectively.
Funding
This work was supported by the National Science and Technology Infrastructure Foundation of China (NIMR-2019); Guizhou Scientific Plan Project [(2019) 2873]; Excellent Youth Talent Training Project of Guizhou Province [(2017) 5639]; Guiyang Science and Technology Project [(2017) No.5–19]; Talent Base Project of Guizhou Province, China [FCJD2018-22]; Research Fund of Education Bureau of Guizhou Province, China [(2018) 481].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare there are no conflicts of interest. The authors have obtained the appropriate permissions from the responsible authorities of Qinghai Oilfield (Tibetan Plateau) for using oil-well production water in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, XJ., Liu, HC., Zhou, YG. et al. Pseudomonas Saliphila sp. nov., a Bacterium Isolated from Oil-Well Production Water in Qinghai Oilfield of China. Curr Microbiol 77, 1924–1931 (2020). https://doi.org/10.1007/s00284-020-01986-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01986-5