Abstract
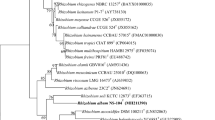
A Gram-stain-negative, facultative aerobic, non-spore-forming, non-motile, non-flagellated, rod-shaped bacterium, designated strain NAU-18T was isolated from an oil-contaminated soil in China. Strain NAU-18T could grow at 10–42 °C (optimum, 30 °C), at pH 5.0–8.0 (optimum, 7.0) and in the presence of 0–2.0% (w/v) NaCl (optimum, 0.5% NaCl in R2A). The predominant fatty acids were C18:1ω7c (71.2%) and Summed feature 2 (5.1%), representing 76.3% of the total fatty acids. The major respiratory quinones were Q9 and Q10. The DNA G + C content of strain NAU-18T was 61.4 mol% based on its draft genome sequence. Genome annotation of strain NAU-18T predicted the presence of 6668 genes, of which 6588 are coding proteins and 80 are RNA genes. Phylogenetic analysis based on 16S rRNA gene sequences revealed that strain NAU-18T was a member of the genus Rhizobium and showed 96.93% (with 93.2% coverage) and 96.81% (with 100% coverage) identities with those of Neorhizobium alkalisoli CCBAU 01393T and Rhizobium oryzicola ZYY136T, respectively. In the phylogenetic analysis, strain NAU-18T and R. oryzicola ZYY136T are consistently placed in the same branch. Strain NAU-18T represents a novel species within the genus Rhizobium, for which the name Rhizobium terrae sp. nov. is proposed, with the type strain NAU-18T (=KCTC 62418T = CCTCC AB 2018075T).



Similar content being viewed by others
References
Lee KB, Te LC, Anzai Y et al (2005) The hierarchical system of the “Alphaproteobacteria”: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol 55:1907–1919. https://doi.org/10.1099/ijs.0.63663-0
Frank B (1889) Ueber die Pilzsymbiose der Leguminosen. Plant Biol 7(8):332–346
Zhang XX, Gao JS, Cao YH et al (2015) Rhizobium oryzicola sp. nov., potential plantgrowth—Promoting endophytic bacteria isolated from rice roots. Int J Syst Evol Microbiol 65:2931–2936. https://doi.org/10.1099/ijs.0.000358
Zhang X-X, Tang X, Sheirdil RA et al (2014) Rhizobium rhizoryzae sp. nov., isolated from rice roots. Int J Syst Evol Microbiol 64:1373–1377. https://doi.org/10.1099/ijs.0.056325-0
Roche P, Lerouge P, Ponthus C, Prome JC (1991) Structural determination of bacterial nodulation factors involved in the Rhizobium meliloti-alfalfa symbiosis. J Biol Chem 266:10933–10940
Gao JL, Sun P, Wang XM et al (2017) Rhizobium wenxiniae sp. nov., an endophytic bacterium isolated from maize root. Int J Syst Evol Microbiol 67:2798–2803. https://doi.org/10.1099/ijsem.0.002025
Afzal A, Bano A (2008) Rhizobium and phosphate solubilizing bacteria improve the yield and phosphorus uptake in wheat (Triticum aestivum). Int J Agric Biol 10:85–88
Peng G, Yuan Q, Li H et al (2008) Rhizobium oryzae sp. nov., isolated from the wild rice Oryza alta. Int J Syst Evol Microbiol 58:2158–2163. https://doi.org/10.1099/ijs.0.65632-0
Puławska J, Willems A, De Meyer SE, Süle S (2012) Rhizobium nepotum sp. nov. isolated from tumors on different plant species. Syst Appl Microbiol 35:215–220. https://doi.org/10.1016/j.syapm.2012.03.001
Wielbo J, Marek-Kozaczuk M, Kidaj D, Skorupska A (2011) Competitiveness of Rhizobium leguminosarum bv. trifolii strains in mixed inoculation of clover (Trifolium pratense). Polish J Microbiol 60:43–49
McInroy JA, Kloepper JW (1995) Population dynamics of endophytic bacteria in field-grown sweet corn and cotton. Can J Microbiol 41:895–901. https://doi.org/10.1139/m95-123
Mousavi SA, Willems A, Nesme X et al (2015) Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90. https://doi.org/10.1016/j.syapm.2014.12.003
Mousavi SA, Österman J, Wahlberg N et al (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215. https://doi.org/10.1016/j.syapm.2013.12.007
An DS, Im WT, Yang HC, Lee ST (2006) Shinella granuli gen. nov., sp. nov., and proposal of the reclassification of Zoogloea ramigera ATCC 19623 as Shinella zoogloeoides sp. nov. Int J Syst Evol Microbiol 56:443–448. https://doi.org/10.1099/ijs.0.63942-0
Kimes NE, López-Pérez M, Flores-Félix JD et al (2015) Pseudorhizobium pelagicum gen. nov., sp. nov. isolated from a pelagic Mediterranean zone. Syst Appl Microbiol 38:293–299. https://doi.org/10.1016/j.syapm.2015.05.003
Tóth E, Szuróczki S, Kéki Z et al (2017) Gellertiella hungarica gen. nov., sp. nov., a novel bacterium of the family Rhizobiaceae isolated from a spa in Budapest. Int J Syst Evol Microbiol 67:4565–4571. https://doi.org/10.1099/ijsem.0.002332
Casida LE (1982) Ensifer adhaerens gen. nov., sp. nov.: a bacterial predator of bacteria in soil. Int J Syst Bacteriol 32:339–345. https://doi.org/10.1099/00207713-32-3-339
Díaz-Cárdenas C, Bernal LF, Caro-Quintero A et al (2017) Draft genome and description of Consotaella salsifontis gen. nov. sp. nov., a halophilic, free-living, nitrogen-fixing alphaproteobacterium isolated from an ancient terrestrial saline spring. Int J Syst Evol Microbiol 67:3744–3751. https://doi.org/10.1099/ijsem.0.002185
Kathiravan R, Jegan S, Ganga V et al (2013) Ciceribacter lividus gen. nov., sp. nov., isolated from rhizosphere soil of chick pea (Cicer arietinum L.). Int J Syst Evol Microbiol 63:4484–4488. https://doi.org/10.1099/ijs.0.049726-0
Tighe SW, De LP, Dipietro K, Lindstro K (2000) Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock Microbial Identification System. Int J Syst Evol Microbiol 50:787–801
Young JM, Kuykendall LD, Martínez-Romero E et al (2001) A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi. Int J Syst Evol Microbiol 51:89–103. https://doi.org/10.1099/00207713-51-1-89
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Lu YL, Chen WF, Han LL et al (2009) Rhizobium alkalisoli sp. nov., isolated from Caragana intermedia growing in saline-alkaline soils in the north of China. Int J Syst Evol Microbiol 59:3006–3011. https://doi.org/10.1099/ijs.0.007237-0
Claus D (1992) A standardized Gram staining procedure. World J Microbiol Biotechnol 8:451–452. https://doi.org/10.1007/BF01198764
Zhang WY, Huo YY, Zhang XQ et al (2013) Halolamina salifodinae sp. nov. and Halolamina salina sp. nov., two extremely halophilic archaea isolated from a salt mine. Int J Syst Evol Microbiol 63:4380–4385. https://doi.org/10.1099/ijs.0.050864-0
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202. https://doi.org/10.1016/0378-1097(90)90282-U
Tindall BJ (1990) A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst Appl Microbiol 13:128–130. https://doi.org/10.1016/S0723-2020(11)80158-X
Tindall BJ, Sikorski J, Smibert RA, Krieg NR (2007) Phenotypic Characterization and the Principles of Comparative Systematics. In: Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, Snyder LR (eds) Methods for general and molecular microbiology, 3rded edn. ASM Pres, Washington, D.C., pp 330–393
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Kuykendall LD, Roy MA, Oneill JJ, Devine TE (1988) Fatty Acids, Antibiotic Resistance, and Deoxyribonucleic Acid Homology Groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361. https://doi.org/10.1099/00207713-38-4-358
Zhang X, Li B, Wang H et al (2012) Rhizobium petrolearium sp. nov., isolated from oilcontaminated soil. Int J Syst Evol Microbiol 62:1871–1876. https://doi.org/10.1099/ijs.0.026880-0
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from micro-organisms. J Mol Biol 3:208–218. https://doi.org/10.1016/S0022-2836(61)80047-8
Du H, Jiao N, Hu Y, Zeng Y (2006) Diversity and distribution of pigmented heterotrophic bacteria in marine environments. FEMS Microbiol Ecol 57:92–105. https://doi.org/10.1111/j.1574-6941.2006.00090.x
Chun J, Lee J-H, Jung Y et al (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261. https://doi.org/10.1099/ijs.0.64915-0
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/BF01731581
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Markowitz VM, Chen IMA, Palaniappan K et al (2014) IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res 42:D560–D567
Pati A, Ivanova NN, Mikhailova N et al (2010) GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. https://doi.org/10.1038/nmeth.1457
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103. https://doi.org/10.1016/S0923-2508(00)01172-4
Sarita S, Sharma PK, Priefer UB, Prell J (2005) Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol Ecol 54:1–11. https://doi.org/10.1016/j.femsec.2005.02.015
De LJ, Cattoir H, Reynaerts A (1970) The Quantitative Measurement of DNA Hybridization from Renaturation Rates. Eur J Biochem. https://doi.org/10.1111/j.1432-1033.1970.tb00830.x
Ebenhöh O, Handorf T, Heinrich R (2004) Structural analysis of expanding metabolic networks. Genome Inform 15:35–45. https://doi.org/10.11234/GI1990.15.35
R Foundation for Statistical Computing (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Boston
Stackebrandt E, Frederiksen W, Garrity GM et al (2002) Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. https://doi.org/10.1099/ijs.0.02360-0
Gevers D, Cohan FM, Lawrence JG et al (2005) Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739
Martens M, Dawyndt P, Coopman R et al (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekee** genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214. https://doi.org/10.1099/ijs.0.65392-0
Martens M, Delaere M, Coopman R et al (2007) Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol 57:489–503. https://doi.org/10.1099/ijs.0.64344-0
Vinuesa P, Silva C, Lorite MJ et al (2005) Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst Appl Microbiol 28:702–716. https://doi.org/10.1016/j.syapm.2005.05.007
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession numbers for the draft genome sequence and 16S rRNA gene, atpD and recA gene sequence of strain NAU-18T are QJKM00000000, MH244125, MH327782 and MH327781, respectively. The annotation information is deposited at the Joint Genomic Institute (JGI), with IMG genome ID 2784132094.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ruan, Zp., Cao, Wm., Zhang, X. et al. Rhizobium terrae sp. nov., Isolated from an Oil-Contaminated Soil in China. Curr Microbiol 77, 1117–1124 (2020). https://doi.org/10.1007/s00284-020-01889-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01889-5