Abstract
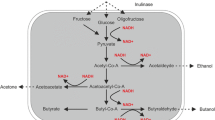
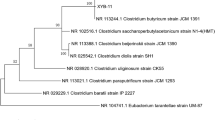
A wild-type solventogenic strain Clostridium diolis WST, isolated from mangrove sediments, was characterized to produce high amount of butanol and acetone with negligible level of ethanol and acids from glucose via a unique acetone-butanol (AB) fermentation pathway. Through the genomic sequencing, the assembled draft genome of strain WST is calculated to be 5.85 Mb with a GC content of 29.69% and contains 5263 genes that contribute to the annotation of 5049 protein-coding sequences. Within these annotated genes, the butanol dehydrogenase gene (bdh) was determined to be in a higher amount from strain WST compared to other Clostridial strains, which is positively related to its high-efficient production of butanol. Therefore, we present a draft genome sequence analysis of strain WST in this article that should facilitate to further understand the solventogenic mechanism of this special microorganism.


Similar content being viewed by others
References
Cooksley CM, Zhang Y, Wang H, Redl S, Winzer K, Minton NP (2012) Targeted mutagenesis of the Clostridium acetobutylicum acetone-butanol-ethanol fermentation pathway. Metab Eng 14(6):630–641
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23(6):673
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L (2008) InterPro: the integrative protein signature database. Nucleic Acids Res 37(S1):D211–D215
Jiang Y, Chen T, Dong W, Zhang M, Zhang W, Wu H, Ma J, Jiang M, **n F (2017) The Draft Genome Sequence of Clostridium beijerinckii NJP7, a unique bacterium capable of producing isopropanol-butanol from hemicellulose through consolidated bioprocessing. Curr Microbiol (3):1–4
Joungmin L, Yusin J, Sungjun C, Jungae I, Song HH, Junghee C, Doyoung S, Papoutsakis ET, Bennett GN, Sangyup L (2012) Metabolic engineering of Clostridium acetobutylicum ATCC 824 for isopropanol-butanol-ethanol fermentation. Appl Environ Microbiol 78(5):1416–1423
Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35(9):3100
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20(2):265–272
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5):955–964
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17(1):10
Minoru K, Michihiro A, Susumu G, Masahiro H, Mika H, Masumi I, Toshiaki K, Shuichi K, Shujiro O, Toshiaki T (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36(Database issue):D480–D484
Sedlar K, Kolek J, Provaznik I, Patakova P (2017) Reclassification of non-type strain Clostridium pasteurianum NRRL B-598 as Clostridium beijerinckii NRRL B-598. J Biotechnol 244:1–3
Shanmugam S, Sun C, Zeng XM, Wu YR (2018) High-efficient production of biobutanol by a novel Clostridium sp. strain WST with uncontrolled pH strategy. Bioresource Technol 256:543–547
Sun C, Zhang S, **n F, Shanmugam S, Wu YR (2018) Genomic comparison of Clostridium species with the potential in utilizing red algal biomass for biobutanol production. Biotechnol Biofuels 11:42
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28(1):33–36
Wu YR, He J (2015) Characterization of a xylanase-producing Cellvibrio mixtus strain J3-8 and its genome analysis. Sci Rep 5:10521
Wu YR, Li Y, Yang KL, He J (2012) Draft genome sequence of butanol-acetone-producing Clostridium beijerinckii strain G117. J Bacteriol 194(19):5470–5471
**n F, Chen T, Jiang Y, Dong W, Zhang W, Zhang M, Wu H, Ma J, Jiang M (2017) Strategies for improved isopropanol-butanol production by a Clostridium strain from glucose and hemicellulose through consolidated bioprocessing. Biotechnol Biofuels 10(1):118
**n F, Dong W, Jiang Y, Ma J, Zhang W, Wu H, Zhang M, Jiang M (2017) Recent advances on conversion and co-production of acetone-butanol-ethanol into high value-added bioproducts. Crit Rev Biotechnol. https://doi.org/10.1080/07388551.2017.1376309
Yang G, Yu J, Hui W, Liu X, Li Z, Jian L, Han X, Shen Z, Dong H, Yang Y (2011) Economical challenges to microbial producers of butanol: Feedstock, butanol ratio and titer. Biotechnol J 6(11):1348
Yi W, Li X, Mao Y, Blaschek HP (2012) Genome-wide dynamic transcriptional profiling in Clostridium beijerinckii NCIMB 8052 using single-nucleotide resolution RNA-SEq. BMC Genomics 13(1):102
Yu W, Fei T, Tang H, ** X (2013) Genome sequence of Clostridium diolis strain DSM 15410, a promising natural producer of 1,3-propanediol. Genome Announc 1(4):e00542-13
Zhao J, Lu C, Chen CC, Yang ST (2013) Biological production of butanol and higher alcohols. Bioprocessing technologies in biorefinery for sustainable production of fuels, chemicals, and polymers. Wiley, Hoboken, pp 235–262
Acknowledgements
This work was financially supported the “Sail Plan” Program for the Introduction of Outstanding Talents of Guangdong Province of China (No. 14600601), the Major University Research Foundation of Guangdong Province of China (No. 2015KQNCX041), the Start-Up Funding of Shantou University (No. NTF15007), the International Cooperation Research Project of Shantou University (No. NC2017001) and the Foundation of Guangdong Provincial Key Laboratory of Marine Biotechnology (No. GPKLMB201702).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared there was no conflict of interest.
Additional information
Nucleotide Sequence Accession Numbers The draft genomic sequence of C. diolis strain WST has been deposited into GenBank with the accession number of PRKY00000000.1.
Rights and permissions
About this article
Cite this article
Chen, C., Sun, C. & Wu, YR. The Draft Genome Sequence of a Novel High-Efficient Butanol-Producing Bacterium Clostridium Diolis Strain WST. Curr Microbiol 75, 1011–1015 (2018). https://doi.org/10.1007/s00284-018-1481-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-018-1481-5