Abstract
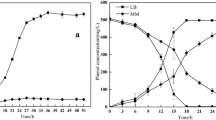
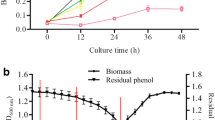
Rhodococcus pyridinivorans B403 is a promising bacterium for degrading phenolic pollutants. In the application, the high-concentration substrate has a significant inhibitory effect on cell growth and phenol degradation, which makes adaptive evolution study of bacteria an important guarantee for further application. The present work found evolved R. pyridinivorans (X1 and X2) had enhanced tolerance to phenolic pollutants as compared to the ancestor strain: the minimum inhibitory concentrations (MIC) of phenol, m-cresol, and catechol increased from 1.2, 0.7, 0.8 g/L to 1.8, 1.0, 1.2 g/L of strain X1, and to 2.4, 1.2, 1.4 g/L of strain X2, respectively. Furthermore, compared to B403, X1, and X2 accumulated more biomass in 500-mg/L cresol medium and degraded phenols more efficiently. Correspondingly, genome sequencing revealed that the mutation sites in genes were annotated as encoding phosphotransferase, MFS transporter, AcrR regulator, and GlpD regulator in the adapted strains, which were closely associated with improved phenol tolerance and degradation. The conclusions provided theoretical basis for the phenol tolerance and degradation, which could promote construction of engineering bacteria for practical application.
Key points
• Evolved strains were more resistant to phenols
• Evolved strains degraded phenols more quickly
• Genome sequencing elucidated mechanisms of enhanced phenol tolerance and degradation






Similar content being viewed by others
Data availability
Statement Raw sequence data of strains B403, X1, and X2 have been deposited in the NCBI Short Read Archive under accession number PRJNA687595, SRX9840402, and SRX9840403.
References
Al-Kandari F, Al-Temaimi R, Van-Vliet AHM, Woodward MJ (2019) Thymol tolerance in Escherichia coli induces morphological, metabolic and genetic changes. BMC Microbiol 19(1):1–11. https://doi.org/10.1186/s12866-019-1663-8
Alsaker KV, Paredes C, Papoutsakis ET (2010) Metabolite stress and tolerance in the production of biofuels and chemicals: gene-expression-based systems analysis of butanol, butyrate, and acetate stresses in the anaerobe Clostridium acetobutylicum. Biotechnol Bioeng 105(6):1131–1147. https://doi.org/10.1002/bit.22628
Cheng D, Li X, Yuan Y, Yang C, Tang T, Zhao Q, Sun Y (2019) Adaptive evolution and carbon dioxide fixation of Chlorella sp. in simulated flue gas. Sci Total Environ 650:2931–2938. https://doi.org/10.1016/j.scitotenv.2018.10.070
Deng W, Li C, **e J (2013) The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cell Signal 25(7):1608–1613. https://doi.org/10.1016/j.cellsig.2013.04.003
Doi Y (2019) Glycerol metabolism and its regulation in lactic acid bacteria. Appl Microbiol Biot 103(13):5079–5093. https://doi.org/10.1007/s00253-019-09830-y
Emelyanova EV, Solyanikova IP (2020) Evaluation of phenol-degradation activity of Rhodococcus opacus 1CP using immobilized and intact cells. Int J Environ Sci Te 17(4):2279–2294. https://doi.org/10.1007/s13762-019-02609-8
Felshia SC, Karthick NA, Thilagam R, Chandralekha A, Raghavarao KSMS, Gnanamani A (2017) Efficacy of free and encapsulated Bacillus lichenformis strain SL10 on degradation of phenol: a comparative study of degradation kinetics. J Environ Manage 197:373–383. https://doi.org/10.1016/j.jenvman.2017.04.005
Feng Y, Lee PH, Wu D, Shih K (2017) Rapid selective circumneutral degradation of phenolic pollutants using peroxymonosulfate–iodide metal-free oxidation: role of iodine atoms. Environ Sci Technol 51(4):2312–2320. https://doi.org/10.1021/acs.est.6b04528
Foo AC, Harvey BG, Metz JJ, Goto NK (2015) Influence of hydrophobic mismatch on the catalytic activity of Escherichia coli GlpG rhomboid protease. Protein Sci 24(4):464–473. https://doi.org/10.1002/pro.2585
Ge B, Liu Y, Liu B, Zhao W, Zhang K (2016) Characterization of novel DeoR-family member from the Streptomyces ahygroscopicus strain CK-15 that acts as a repressor of morphological development. Appl Microbiol Biot 100(20):8819–8828. https://doi.org/10.1007/s00253-016-7661-y
Jeckelmann JM, Erni B (2020) Transporters of glucose and other carbohydrates in bacteria. Pflüg Arch Eur J Phy 472(9):1129–1153. https://doi.org/10.1007/s00424-020-02379-0
Jiang Y, Shang Y, Yang K, Wang H (2016) Phenol degradation by halophilic fungal isolate JS4 and evaluation of its tolerance of heavy metals. Appl Microbiol Biot 100(4):1883–1890. https://doi.org/10.1007/s00253-015-7180-2
Kazuhiko K, Josephine L, Anthony JS (2015) Tolerance and adaptive evolution of triacylglycerol-producing Rhodococcus opacus to lignocellulose-derived inhibitors. Biotechnol Biofuels 8(1):1–14. https://doi.org/10.1186/s13068-015-0258-3
Kim J, Kim J (2014) Arsenite oxidation-enhanced photocatalytic degradation of phenolic pollutants on platinized TiO2. Environ Sci Technol 48(22):13384–13391. https://doi.org/10.1021/es504082r
Klinke HB, Thomsen AB, Ahring BK (2004) Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biot 66(1):10–26. https://doi.org/10.1007/s00253-004-1642-2
Kowalska-Krochmal B, Dudek-Wicher R (2021) The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens 10(2):165. https://doi.org/10.3390/pathogens10020165
Kumar V, Singh B, Van-Belkum MJ, Diep DB, Chikindas ML, Ermakov AM, Tiwari SK (2021) Halocins, natural antimicrobials of archaea: exotic or special or both? Biotechnol Adv 53:107834. https://doi.org/10.1016/j.biotechadv.2061.107834
LaCroix RA, Palsson BO, Feist AM (2017) A model for designing adaptive laboratory evolution experiments. Appl Environ Microb 83(8):e03115-e3116. https://doi.org/10.1128/AEM.03115-16
Lefebvre O, Moletta R (2006) Treatment of organic pollution in industrial saline wastewater: a literature review. Water Res 40(20):3671–3682. https://doi.org/10.1016/j.watres.2006.08.027
Li CM, Wu HZ, Wang YX, Zhu S, Wei CH (2020) Enhancement of phenol biodegradation: metabolic division of labor in co-culture of Stenotrophomonas sp. N5 and Advenella sp. B9. J Hazard Mater 400:123214. https://doi.org/10.1016/j.jhazmat.2020.123214
Li D, Wang L, Zhao Q, Wei W, Sun Y (2015) Improving high carbon dioxide tolerance and carbon dioxide fixation capability of Chlorella sp. by adaptive laboratory evolution. Bioresource Technol 185:269–275. https://doi.org/10.1016/j.biortech.2015.03.011
Li X, Pei G, Liu L, Chen L, Zhang W (2017) Metabolomic analysis and lipid accumulation in a glucose tolerant Crypthecodinium cohnii strain obtained by adaptive laboratory evolution. Bioresource Technol 235:87–95. https://doi.org/10.1016/j.biortech.2017.03.049
Li Y, Li J, Wang C, Wang P (2010) Growth kinetics and phenol biodegradation of psychrotrophic Pseudomonas putida LY1. Bioresource Technol 101(17):6740–6744. https://doi.org/10.1016/j.biortech.2010.03.083
Lin J, Sharma V, Milase R, Mbhense N (2016) Simultaneous enhancement of phenolic compound degradations by Acinetobacter strain V2 via a step-wise continuous acclimation process. J Basic Microb 56(6):627–634. https://doi.org/10.1002/jobm.201500263
Martínez-Gómez K, Flores N, Castañeda HM, Martínez-Batallar G, Hernández-Chávez G, Ramírez OT, Gosset G, Encarnación S, Bolivar F (2012) New insights into Escherichia coli metabolism: carbon scavenging, acetate metabolism and carbon recycling responses during growth on glycerol. Microb Cell Fact 11(1):1–21. https://doi.org/10.1186/1475-2859-11-46
Matarredona L, Camacho M, Zafrilla B, Bonete MJ, Esclapez J (2020) The role of stress proteins in haloarchaea and their adaptive response to environmental shifts. Biomolecules 10(10):1390. https://doi.org/10.3390/biom10101390
Nazos TT, Kokarakis E, Ghanotakis DF (2017) Metabolism of xenobiotics by Chlamydomonas reinhardtii: phenol degradation under conditions affecting photosynthesis. Photosynth Res 131(1):31–40. https://doi.org/10.1007/s11120-016-0294-2
Nuonming P, Khemthong S, Dokpikul T, Sukchawalit R, Mongkolsuk S (2018) Characterization and regulation of AcrABR, a RND-type multidrug efflux system, in Agrobacterium tumefaciens C58. Microbiol Res 214:146–155. https://doi.org/10.1016/j.micres.2018.06.014
Oide S, Gunji W, Moteki Y, Yamamoto S, Suda M, Jojima T, Yukawa H, Inui M (2015) Thermal and solvent stress cross-tolerance conferred to Corynebacterium glutamicum by adaptive laboratory evolution. Appl and Environ Microb 81(7):2284–2298. https://doi.org/10.1128/AEM.03973-14
Pajuelo RD, Čunátová K, Vrbacký M, Pecinová A, Houštěk J, Mráček T, Pecina P (2020) Cytochrome c oxidase subunit 4 isoform exchange results in modulation of oxygen affinity. Cells 9(2):443. https://doi.org/10.3390/cells9020443
Pasqua M, Grossi M, Zennaro A, Fanelli G, Micheli G, Barras F, Colonna B, Prosseda G (2019) The varied role of efflux pumps of the MFS family in the interplay of bacteria with animal and plant cells. Microorganisms 7(9):285. https://doi.org/10.3390/microorganisms7090285
Pitceathly RD, Rahman S, Wedatilake Y, Polke JM, Cirak S, Foley AR, Sailer A, Hurles ME, Matthew E, Stalker J, Hargreaves I, Woodward CE, Sweeney MG, Muntoni F, Houlden H, Taanman JW, Hanna M (2013) NDUFA4 mutations underlie dysfunction of a cytochrome c oxidase subunit linked to human neurological disease. Cell Rep 3(6):1795–1805. https://doi.org/10.1016/j.celrep.2013.05.005
Roell GW, Carr RR, Campbell T, Shang Z, Henson WR, Czajka JJ, Garcia MH, Zhang F, Foston M, Dantas G, Moon TS, Tang YJ (2019) A concerted systems biology analysis of phenol metabolism in Rhodococcus opacus PD630. Metab Eng 55:120–130. https://doi.org/10.1016/j.ymben.2019.06.013
Ruan B, Wu P, Chen M, Lai X, Chen L, Yu L, Gong B, Kang C, Dang Z, Shi Z, Liu Z (2018) Immobilization of Sphingomonas sp. GY2B in polyvinyl alcohol–alginate–kaolin beads for efficient degradation of phenol against unfavorable environmental factors. Ecotox Environ Safe 162:103–111. https://doi.org/10.1016/j.ecoenv.2018.06.058
Shui Z, Qin H, Wu B, Ruan Z, Wang L, Tan F, Wang J, Tang X, Dai L, Hu G, He M (2015) Adaptive laboratory evolution of ethanologenic Zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl Microbiol and Biot 99(13):5739–5748. https://doi.org/10.1007/s00253-015-6616-z
Subhadra B, Surendran S, Kim DH, Woo K, Oh MH, Choi CH (2019) The transcription factor NemR is an electrophile-sensing regulator important for the detoxification of reactive electrophiles in Acinetobacter nosocomialis. Res Microbiol 170(3):123–130. https://doi.org/10.1016/j.resmic.2019.02.001
Tagel M, Ilves H, Leppik M, Jürgenstein K, Remme J, Kivisaar M (2021) Pseudouridines of tRNA anticodon stem-loop have unexpected role in mutagenesis in Pseudomonas sp. Microorganisms 9:25. https://doi.org/10.3390/microorganisms9010025
Van-Houdt R, Vandecraen J, Leys N, Monsieurs P, Aertsen A (2021) Adaptation of Cupriavidus metallidurans CH34 to toxic zinc concentrations involves an uncharacterized ABC-Type transporter. Microorganisms 9(2):309. https://doi.org/10.3390/microorganisms9020309
Wang L, Xue C, Wang L, Zhao Q, Wei W, Sun Y (2016) Strain improvement of Chlorella sp. for phenol biodegradation by adaptive laboratory evolution. Bioresource Ttechnol 205:264–268. https://doi.org/10.1016/j.biortech.2016.01.022
Warner JB, Lolkema JS (2003) CcpA-dependent carbon catabolite repression in bacteria. Microbiol Mol Biol R 67(4):475–490. https://doi.org/10.1128/MMBR.67.4.475-490.2003
Wu S, Liu H, Lin Y, Yang C, Lou W, Sun J, Du C, Zhang D, Nie L, Yin K, Zhong Y (2020) Insights into mechanisms of UV/ferrate oxidation for degradation of phenolic pollutants: role of superoxide radicals. Chemosphere 244:125490. https://doi.org/10.1016/j.chemosphere.2019.125490
**e X, Liu J, Jiang Z, Li H, Ye M, Pan H, Zhu J, Song H (2021) The conversion of the nutrient condition alter the phenol degradation pathway by Rhodococcus biphenylivorans B403: a comparative transcriptomic and proteomic approach. Environ Sci Pollut R 28:56152–56163. https://doi.org/10.1007/s11356-021-14374-8
Xu K, Lee YS, Li J, Li C (2019) Resistance mechanisms and reprogramming of microorganisms for efficient biorefinery under multiple environmental stresses. Synth Syst Biotechnol 4(2):92–98. https://doi.org/10.1016/j.synbio.2019.02.003
Yan N, An M, Chu J, Cao L, Zhu G, Wu W, Wang L, Zhang Y, Rittmann BE (2021) More rapid dechlorination of 2, 4-dichlorophenol using acclimated bacteria. Bioresource Technol 326:124738. https://doi.org/10.1016/j.biortech.2021.124738
Yang E, Fan L, Yan J, Jiang Y, Doucette C, Fillmore S, Walker B (2018) Influence of culture media, pH and temperature on growth and bacteriocin production of bacteriocinogenic lactic acid bacteria. AMB Express 8(1):1–14. https://doi.org/10.1186/s13568-018-0536-0
Yoneda A, Henson WR, Goldner NK, Park KJ, Forsberg KJ, Kim SJ, Pesesky MW, Foston M, Dantas G, Moon TS (2016) Comparative transcriptomics elucidates adaptive phenol tolerance and utilization in lipid-accumulating Rhodococcus opacus PD630. Nucleic Acids Res 44(5):2240–2254. https://doi.org/10.1093/nar/gkw055
Zeng L, Burne RA (2018) Essential roles of the sppRA fructose-phosphate phosphohydrolase operon in carbohydrate metabolism and virulence expression by streptococcus mutans. J Bacteriol 201(2):e00586-e618. https://doi.org/10.1128/JB.00586-18
Zhou L, Xu Z, Wen Z, Lu M, Wang Z, Zhang Y, Zhou H, ** M (2021) Combined adaptive evolution and transcriptomic profiles reveal aromatic aldehydes tolerance mechanisms in Yarrowia lipolytica. Bioresource Technol 329:124910. https://doi.org/10.1016/j.biortech.2021.124910
Acknowledgements
Authors would like to thank Editage (www.editage.cn) for English language editing.
Funding
This work was supported by China National Key R&D Program (2020YFA0908400; 2019YFA0905000), the Science and Technology Innovation Program of Hubei Province (2020BBA056; 2019ABA117), the Central Committee Guides Local Science and Technology Development Projects (2018ZYYD034), and the Wuhan Science and Technology Plan (2019020701011496).
Author information
Authors and Affiliations
Contributions
Conceptualization: H. T. Song and Z. B. Jiang.; methodology: M. Ye and Z. B. Jiang; software: F. Peng; validation: M. Ye; formal analysis: M.Y e, T. Y. Zhang, A. H. Luo; investigation: M. Ye; resources: M. Ye; data curation: M. Ye and F. Peng; writing—original draft preparation: M. Ye and F. Peng; writing—review and editing: F. Peng and H. T. Song; visualization: M. Ye and F. Peng; supervision: J. S. Liu Y. F. Liu and Y. Lan; project administration: Z. B. Jiang; funding acquisition: H. T. Song and Z. B. Jiang. All the authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Peng, F., Ye, M., Liu, Y. et al. Comparative genomics reveals response of Rhodococcus pyridinivorans B403 to phenol after evolution. Appl Microbiol Biotechnol 106, 2751–2761 (2022). https://doi.org/10.1007/s00253-022-11858-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11858-6