Abstract
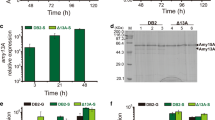
Filamentous fungi are extensively used as an important expression host for the production of a variety of essential industrial proteins. They have significant promise as an expression system for protein synthesis due to their inherent superior secretory capabilities. The purpose of this study was to develop a novel expression system by utilizing a Penicillium oxalicum strain that possesses a high capacity for protein secretion. The expression of glycoside hydrolases in P. oxalicum was evaluated in a cleaner extracellular background where the formation of two major amylases was inhibited. Four glycoside hydrolases (CBHI, Amy15B, BGL1, and Cel12A) were expressed under the highly constitutive promoter PubiD. It was found that the proteins exhibited high purity in the culture supernatant after cultivation with starch. Two inducible promoters, Pamy15A and PempA, under the activation of the transcription factor AmyR were used as elements in the construction of versatile vectors. When using the cellobiohydrolase CBHI as the extracellular quantitative reporter, the empA promoter screened from the AmyR-overexpressing strain was shown to be superior to the amy15A promoter based on RNA-sequencing data. Therefore, we designed an expression system consisting of a cleaner background host strain and an adjustable promoter. This system enables rapid and high-throughput evaluation of glycoside hydrolases from filamentous fungi.
Key points
• A new protein expression system derived from Penicillium oxalicum has been developed.
• The expression platform is capable of secreting recombinant proteins with high purity.
• The adjustable promoter may allow for further optimization of recombinant protein synthesis.








Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary files.
References
Ahmad M, Hirz M, Pichler H, Schwab H (2014) Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Appl Microbiol Biotechnol 98(12):5301–5317. https://doi.org/10.1007/s00253-014-5732-5
Boer H, Teeri TT, Koivula A (2000) Characterization of Trichoderma reesei cellobiohydrolase Cel7A secreted from Pichia pastoris using two different promoters. Biotechnol Bioeng 69(5):486–494. https://doi.org/10.1002/1097-0290(20000905)69:5%3c486::aid-bit3%3e3.0.co;2-n
Chokhawala HA, Roche CM, Kim TW, Atreya ME, Vegesna N, Dana CM, Blanch HW, Clark DSJBB (2015) Mutagenesis of Trichoderma reesei endoglucanase I: impact of expression host on activity and stability at elevated temperatures. BMC Biotechnol 15(1). https://doi.org/10.1186/s12896-015-0118-z
Fleissner A, Dersch P (2010) Expression and export: recombinant protein production systems for Aspergillus. Appl Microbiol Biotechnol 87(4):1255–1270. https://doi.org/10.1007/s00253-010-2672-6
Gao L, Li Z, **a C, Qu Y, Liu M, Yang P, Yu L, Song X (2017) Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol Biofuels 10(1):100. https://doi.org/10.1186/s13068-017-0783-3
Gerngross TU (2004) Advances in the production of human therapeutic proteins in yeasts and filamentous fungi. Nat Biotechnol 22(11):1409–1414. https://doi.org/10.1038/nbt1028
Gressler M, Hortschansky P, Geib E, Brock M (2015) A new high-performance heterologous fungal expression system based on regulatory elements from the Aspergillus terreus terrein gene cluster. Front Microbiol 6:184. https://doi.org/10.3389/fmicb.2015.00184
Hamilton SR, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, Wildt S, Gerngross TU (2003) Production of complex human glycoproteins in yeast. Science 301(5637):1244–1246. https://doi.org/10.1126/science.1088166
Han L, Liu K, Ma W, Jiang Y, Hou S, Tan Y, Yuan Q, Niu K, Fang X (2020) Redesigning transcription factor Cre1 for alleviating carbon catabolite repression in Trichoderma reesei. Synth Syst Biotechnol 5(3):230–235. https://doi.org/10.1016/j.synbio.2020.07.002
Hu Y, Liu G, Li Z, Qin Y, Qu Y, Song X (2013) G protein-cAMP signaling pathway mediated by PGA3 plays different roles in regulating the expressions of amylases and cellulases in Penicillium decumbens. Fungal Genet Biol 58–59:62–70. https://doi.org/10.1016/j.fgb.2013.08.002
Hu Y, Xue H, Liu G, Song X, Qu Y (2015) Efficient production and evaluation of lignocellulolytic enzymes using a constitutive protein expression system in Penicillium oxalicum. J Ind Microbiol Biotechnol 42(6):877–887. https://doi.org/10.1007/s10295-015-1607-8
Li C, Zhou J, Du G, Chen J, Takahashi S, Liu S (2020) Develo** Aspergillus niger as a cell factory for food enzyme production. Biotechnol Adv 44:107630. https://doi.org/10.1016/j.biotechadv.2020.107630
Li D, Bo Z, Li S, Jie Z, Cao H, Huang Y, Cui Z (2017) A novel vector for construction of markerless multicopy overexpression transformants in Pichia pastoris. Front Microbiol 8:1698. https://doi.org/10.3389/fmicb.2017.01698
Li Z, Yao G, Wu R, Gao L, Kan Q, Liu M, Yang P, Liu G, Qin Y, Song X (2015) Synergistic and dose-controlled regulation of cellulase gene expression in Penicillium oxalicum. PLoS Genet 11(9):e1005509. https://doi.org/10.1371/journal.pgen.1005509
Li ZH, Du CM, Zhong YH, Wang TH (2010) Development of a highly efficient gene targeting system allowing rapid genetic manipulations in Penicillium decumbens. Appl Microbiol Biotechnol 87(3):1065. https://doi.org/10.1007/s00253-010-2566-7
Linger JG, Taylor LE, Baker JO, Vander Wall T, Hobdey SE, Podkaminer K, Himmel ME, Decker SR (2015) A constitutive expression system for glycosyl hydrolase family 7 cellobiohydrolases in Hypocrea jecorina. Biotechnol Biofuels 8:45. https://doi.org/10.1186/s13068-015-0230-2
Liu G, Zhang L, Wei X, Zou G, Qin Y, Ma L, Li J, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Zhou Z, Qu Y (2013b) Genomic and secretomic analyses reveal unique features of the lignocellulolytic enzyme system of Penicillium decumbens. PLoS One 8(2):e55185. https://doi.org/10.1371/journal.pone.0055185
Liu G, Zhang L, Qin Y, Zou G, Li Z, Yan X, Wei X, Chen M, Chen L, Zheng K, Zhang J, Ma L, Li J, Liu R, Xu H, Bao X, Fang X, Wang L, Zhong Y, Liu W, Zheng H, Wang S, Wang C, Xun L, Zhao GP, Wang T, Zhou Z, Qu Y (2013a) Long-term strain improvements accumulate mutations in regulatory elements responsible for hyper-production of cellulolytic enzymes. Sci Rep 3:1569. https://doi.org/10.1038/srep01569
Lv X, Zheng F, Li C, Zhang W, Chen G, Liu W (2015) Characterization of a copper responsive promoter and its mediated overexpression of the xylanase regulator 1 results in an induction-independent production of cellulases in Trichoderma reesei. Biotechnol Biofuels 8:67. https://doi.org/10.1186/s13068-015-0249-4
Madhavan A, Pandey A, Sukumaran RK (2017) Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis. Bioresour Technol 245(Pt B):1334–1342. https://doi.org/10.1016/j.biortech.2017.05.140
Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EG, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, **e G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26(5):553–60. https://doi.org/10.1038/nbt1403
Mattanovich D, Branduardi P, Dato L, Gasser B, Sauer M, Porro D (2012) Recombinant protein production in yeasts. Methods Mol Biol 824:329–358. https://doi.org/10.1007/978-1-61779-433-9_17
Meng QS, Zhang F, Wang W, Liu CG, Zhao XQ, Bai FW (2020) Engineering the effector domain of the artificial transcription factor to improve cellulase production by Trichoderma reesei. Front Bioeng Biotechnol 8:675. https://doi.org/10.3389/fbioe.2020.00675
Nakayashiki H, Hanada S, Nguyen BQ, Kadotani N, Tosa Y, Mayama S (2005) RNA silencing as a tool for exploring gene function in ascomycete fungi. Fungal Genet Biol 42(4):275–283. https://doi.org/10.1016/j.fgb.2005.01.002
Nakazawa H, Kawai T, Ida N, Shida Y, Ogasawara W (2011) Construction of a recombinant Trichoderma reesei strain expressing Aspergillus aculeatus β-glucosidase 1 for efficient biomass conversion. Biotechnol Bioeng 109(1):92–99. https://doi.org/10.1002/bit.23296
Peterson R, Nevalainen H (2011) Trichoderma reesei RUT-C30-thirty years of strain improvement. Microbiology 158(Pt 1):58–68. https://doi.org/10.1099/mic.0.054031-0
Pi C, Zhang Z, **ang B, Tian H, Hu S (2020) Constructing a novel expression system by specific activation of amylase expression pathway in Penicillium. Microb Cell Fact 19(1). https://doi.org/10.1186/s12934-020-01410-4
Satoshi W, Nanami N, Chiaki O, Hiroko T, Yoji H (2018) Modified expression of multi-cellulases in a filamentous fungus Aspergillus oryzae. Bioresour Technol 276:146–153. https://doi.org/10.1016/j.biortech.2018.12.117
Singh A, Taylor LE 2nd, Vander Wall TA, Linger J, Himmel ME, Podkaminer K, Adney WS, Decker SR (2015) Heterologous protein expression in Hypocrea jecorina: a historical perspective and new developments. Biotechnol Adv 33(1):142–154. https://doi.org/10.1016/j.biotechadv.2014.11.009
Tamayo EN, Villanueva A, Hasper AA, de Graaff LH, Ramón D, Orejas M (2008) CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol 45(6):984–993. https://doi.org/10.1016/j.fgb.2008.03.002
Tani S, Itoh T, Kato M, Kobayashi T, Tsukagoshi N (2001) In vivo and in vitro analyses of the AmyR binding site of the Aspergillus nidulans agdA promoter; requirement of the CGG direct repeat for induction and high affinity binding of AmyR. Biosci Biotechnol Biochem 65(7):1568–1574. https://doi.org/10.1271/bbb.65.1568
Tsang A, Butler G, Powlowski J, Panisko EA, Baker SE (2009) Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet Biol 46(1-supp-S):S153–S160. https://doi.org/10.1016/j.fgb.2008.07.014
Uzbas F, Sezerman U, Hartl L, Kubicek CP, Seiboth B (2012) A homologous production system for Trichoderma reesei secreted proteins in a cellulase-free background. Appl Microbiol Biotechnol 93(4):1601–1608. https://doi.org/10.1007/s00253-011-3674-8
Vogel HJ (1956) A convenient growth medium for Neurospora crassa. Microbiol Genet Bull 13:42–43
Ward OP (2012) Production of recombinant proteins by filamentous fungi. Biotechnol Adv 30(5):1119–1139. https://doi.org/10.1016/j.biotechadv.2011.09.012
**a C, Li Z, Xu Y, Yang P, Gao L, Yan Q, Li S, Wang Y, Qu Y, Song X (2019) Introduction of heterologous transcription factors and their target genes into Penicillium oxalicum leads to increased lignocellulolytic enzyme production. Appl Microbiol Biotechnol 103(6):2675–2687. https://doi.org/10.1007/s00253-018-09612-y
Zhang H, Wang S, Zhang XX, Ji W, Song FP, Zhao Y, Li J (2016) The amyR-deletion strain of Aspergillus niger CICC2462 is a suitable host strain to express secreted protein with a low background. Microb Cell Fact 15(1):68. https://doi.org/10.1186/s12934-016-0463-1
Zou G, Shi S, Jiang Y, van den Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang C, Zhou Z (2012) Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Fact 11(1):21. https://doi.org/10.1186/1475-2859-11-21
Acknowledgements
We thank Prof. Yinbo Qu of the State Key Laboratory of Microbial Technology, for providing Penicillium oxalicum strains and pyrG marker recycling plasmid.
Funding
This study was funded by National Natural Science Foundation of China (No. 31700019), the Research Foundation of Education Bureau of Hunan Province (No. 17K058), Hunan province college students research learning and innovative experiment project (No. S202110542106), and the Natural Science Foundation of Hunan Province (No. 2017JJ3203).
Author information
Authors and Affiliations
Contributions
ZZ carried out the experiments and drafted the manuscript. XBY, ZSF, and YL participated in the design of the study. CY and HSB supervised the experiments and helped to draft the manuscript. HYB conceived the study and reviewed the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Z., **ang, B., Zhao, S. et al. Construction of a novel filamentous fungal protein expression system based on redesigning of regulatory elements. Appl Microbiol Biotechnol 106, 647–661 (2022). https://doi.org/10.1007/s00253-022-11761-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11761-0