Abstract
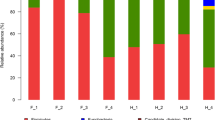
The gut microbiome is integral for the host’s living and environmental adaptation and crucially important for understanding host adaptive mechanisms. The red fox (Vulpes vulpes) dominates a wider ecological niche and more complicated habitat than that of the corsac fox (V. corsac). However, the adaptive mechanisms (in particular, the gut microbiome responsible for this kind of difference) are still unclear. Therefore, we investigated the gut microbiome of these two species in the Hulunbuir grassland, China, and evaluated their microbiome composition, function, and adaptive mechanisms. We profiled the gut microbiome and metabolism function of red and corsac foxes via 16S rRNA gene and metagenome sequencing. The foxes harbored species-specific microbiomes and functions that were related to ecological niche and habitat. The red fox had abundant Bacteroides, which leads to significant enrichment of metabolic pathways (K12373 and K21572) and enzymes related to chitin and carbohydrate degradation that may help the red fox adapt to a wider niche. The corsac fox harbored large proportions of Blautia, Terrisporobacter, and ATP-binding cassette (ABC) transporters (K01990, K02003, and K06147) that can help maintain corsac fox health, allowing it to live in harsh habitats. These results indicate that the gut microbiome of the red and corsac foxes may have different abilities which may provide these species with differing capabilities to adapt to different ecological niches and habitats, thus providing important microbiome data for understanding the mechanisms of host adaptation to different niches and habitats.







Similar content being viewed by others
Data Availability
1. The species identification data can be found in the Genbank of NCBI, accession numbers: CH1 (MT795165.1), CH2 (MT795166.1), CH3 (MT795167.1), CH4 (MT795168.1), CH5 (MT795169.1), CH7 (MT795170.1), CH8 (MT795171.1), CH9 (MT795172.1), CH11 (MT795173.1), CH13 (MT795174.1), SH14 (MT795175.1), SH15 (MT795178.1), SH16 (MT795176.1), SH17 (MT795177.1), and SH21 (MT795179.1).
2. The 16S rRNA gene sequencing data can be found in the NCBI Sequence Read Archive database (accession number PRJNA646772).
3. The metagenomics sequencing data can be found in the NCBI Sequence Read Archive database (accession number PRJNA646811).
Code Availability (Software Application or Custom Code)
Applicable.
References
Greene LK, Williams CV, Junge RE, Mahefarisoa KL, Rajaonarivelo T, Rakotondrainibe H, O’Connell TM, Drea CM (2020) A role for gut microbiota in host niche differentiation. ISME J 14:1675–1687. https://doi.org/10.1038/s41396-020-0640-4
Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, Zhang X, Huang H, Bridgewater LC, Jiang Y (2015) The Bamboo-Eating Giant Panda Harbors a Carnivore-Like Gut Microbiota, with Excessive Seasonal Variations. MBio 6(3):e00022–15
Wu Q, Wang X, Ding Y, Hu Y, Nie Y, Wei W, Ma S, Yan L, Zhu L, Wei F (2017) Seasonal variation in nutrient utilization shapes gut microbiome structure and function in wild giant pandas. Proc R Soc B Biol Sci 284:20170955
Wei F, Wang X, Wu Q (2015) The giant panda gut microbiome. Trends Microbiol 23:450–452
Roggenbuck M, Schnell IB, Blom N, Baelum J, Bertelsen MF, Sicheritzponten T, Sorensen SJ, Gilbert MTP, Graves GR, Hansen LH (2014) The microbiome of New World vultures. Nat Commun 5:5498
Lyu T, Liu G, Zhang H, Wang L, Zhang H (2018) Changes in feeding habits promoted the differentiation of the composition and function of gut microbiotas between domestic dogs (Canis lupus familiaris) and gray wolves (Canis lupus). AMB Express 8(1):1–12
Swanson KS (2016) 0226 Dietary manipulation of canine and feline gut microbiome. J Anim Sci 94:107–107
Li K, Dan Z, Gesang L, Wang H, Zhou Y, Du Y, Ren Y, Shi Y, Nie Y (2016) Comparative analysis of gut microbiota of native Tibetan and Han populations living at different altitudes. PloS one 11:e0155863
Sun G, Zhang H, Wei Q, Zhao C, Yang X, Wu X, **a T, Liu G, Zhang L, Gao Y (2019) Comparative analyses of fecal microbiota in European mouflon (Ovis orientalis musimon) and blue sheep (Pseudois nayaur) living at low or high altitudes. Front Microbiol 10:1735
Zhang Z, Xu D, Wang L, Hao J, Wang J, Zhou X, Wang W, Qiu Q, Huang X, Zhou J (2016) Convergent evolution of rumen microbiome in high-altitude mammals. Curr Biol 26:1873–1879
Bo T, Zhang X, Wen J, Deng K, Qin X, Wang D (2019) The microbiota-gut-brain interaction in regulating host metabolic adaptation to cold in male Brandt’s voles (Lasiopodomys brandtii). ISME J 13:1–17
Schmidt E, Mykytczuk N, Schulte-Hostedde AI (2019) Effects of the captive and wild environment on diversity of the gut microbiome of deer mice (Peromyscus maniculatus). ISME J 13(5):1293–1305
Donadio E, Buskirk SW (2006) Diet, morphology, and interspecific killing in Carnivora. Am Nat 167:524–536
Murdoch JD, Munkhzul T, Buyandelger S, Reading RP, Sillero-Zubiri C (2010) Seasonal food habits of corsac and red foxes in Mongolia and the potential for competition. Mamm Biol 75(1):36–44
Schoener et al (1974) Resource partitioning in ecological communities. Science 185:27–39
De Leon LF, Podos J, Gardezi T, Herrel A, Hendry AP (2014) Darwin’s finches and their diet niches: the sympatric coexistence of imperfect generalists. J Evol Biol 27:1093–1104
Kartzinel TR, Chen P, Coverdale TC, Erickson DL, Kress WJ, Kuzmina M, Rubenstein DI, Wang W, Pringle RM (2015) DNA metabarcoding illuminates dietary niche partitioning by African large herbivores. Proc Natl Acad Sci USA 112:8019–8024
Munkhzul T, Murdoch JD, Reading RP (2012) Home range characteristics of corsac and red foxes in Mongolia. University of Nebraska. https://digitalcommons.unl.edu/biolmongol/12
Saiful AA, Idris AH, Rashid YN, Tamura N, Hayashi F (2001) Home range size of sympatric squirrel species inhabiting a lowland dipterocarp forest in Malaysia. Biotropica 33:346–351
Murdoch JD, Munkhzul T, Buyandelger S, Reading RP (2009) Body size and sexual dimorphism among a population of corsac and red foxes in central Mongolia. Mammalia 73(1):72–75
张洪海, 窦华山, 翟红昌, 吴牧仁 (2006) 三种犬科动物春季洞穴特征. 生态学报: 3980–3988. http://www.ecologica.cn/stxb/ch/reader/view_abstract.aspx?file_no=1000-0933200612-3980-09
Lalitha S (2000) Primer Premier 5. Biotech Softw Internet Rep 1:270–272
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Singh A, Rai A, Tripathi G, Nand V, Gupta N, Srivastava K (2019) Development of species-specific polymerase chain reaction (PCR) targeting on mitochondrial D-loop for identification of buffalo and goat raw meat. J Biol Eng Res Rev 6:01–04
Anabalón L, Encina-Montoya F, Sánchez P, Solano J, Benavente F, Guiñez B, Olivares F, Oberti C, Vega R (2019) High-resolution melting of the cytochrome B gene in fecal DNA: a powerful approach for fox species identification of the Lycalopex genus in Chile. Ecol Evol 9:7448–7454
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Karschmizrachi I, Ouellette BF (1998) The GenBank sequence database. Methods Biochem Anal 39:16
Wu X, Zhang H, Chen J, Shang S, Yan J, Chen Y, Tang X, Zhang H (2017) Analysis and comparison of the wolf microbiome under different environmental factors using three different data of Next Generation Sequencing. Sci Rep 7:11332
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. https://doi.org/10.1038/nmeth.f.303
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21:494–504
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. https://doi.org/10.1128/AEM.00062-07
Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5:299–314
White JR, Nagarajan N, Pop M (2009) Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:e1000352
Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, Nielsen J, Backhed F (2013) Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103
Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, Backhed F, Nielsen J (2012) Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 3:1245–1245
Luo R, Liu B, **e Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1:30–30
Kanehisa M, Goto S, Hattori M, Aokikinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M (2006) From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res 34:354–357
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B (2009) The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:233–238
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Wu X, Zhang H, Chen J, Shang S, Wei Q, Yan J, Tu X (2016) Comparison of the fecal microbiota of dholes high-throughput Illumina sequencing of the V3–V4 region of the 16S rRNA gene. Appl Microbiol Biotechnol 100:3577–3586. https://doi.org/10.1007/s00253-015-7257-y
Wu X, Shang Y, Wei Q, Chen J, Zhang H, Chen Y, Gao X, Wang Z, Zhang H (2020) Gut microbiota in dholes during estrus. Front Microbiol 11:3044. https://doi.org/10.3389/fmicb.2020.575731
Rajilicstojanovic M, De Vos WM (2014) The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38:996–1047
Cummings JH (1981) Short chain fatty acids in the human colon. Gut 22:763–779
Bloemen JG, Venema K, De Poll MCGV, Damink SWMO, Buurman WA, Dejong CHC (2009) Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin Nutr 28:657–661
Flint HJ, Scott KP, Duncan SH, Louis P, Forano E (2012) Microbial degradation of complex carbohydrates in the gut. Gut microbes 3:289–306
Fuerst JA, Sagulenko E (2011) Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol 9:403–413. https://doi.org/10.1038/nrmicro2578
Munoztamayo R, Laroche B, Walter E, Dore J, Duncan SH, Flint HJ, Leclerc M (2011) Kinetic modelling of lactate utilization and butyrate production by key human colonic bacterial species. FEMS Microbiol Ecol 76:615–624
Cook SI, Sellin JH (1998) Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther 12:499–507
Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey FE, Wang T (2013) Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 110:4410–4415
Lin HV, Frassetto A, Kowalik EJ, Nawrocki AR, Lu MM, Kosinski JR, Hubert J, Szeto D, Yao X, Forrest G (2012) Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PloS One 7(4):e35240
Clarke VA, Platt N, Butters TD (1995) Cloning and expression of the β-N-acetylglucosaminidase gene from Streptococcus pneumoniae: generation of truncated enzymes with modified aglycon specificity. J Biol Chem 270(15):8805–8814
Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly J, Parsons J, Dalmolin CGDO, Palfreyman RW, Nielsen LK, Cooper ME (2016) Genomic characterization of the uncultured Bacteroidales family S24–7 inhabiting the guts of homeothermic animals. Microbiome 4:36–36
Reeves AR, Wang GR, Salyers AA (1997) Reeves AR, Wang G-R, Salyers AA. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol 179: 643–649. J Bacteriol 179:643–649
Tuson HH, Foley MH, Koropatkin NM, Biteen JS (2018) The starch utilization system assembles around stationary starch-binding proteins. Biophys J 115(2):242–250
Mcnulty NP, Wu M, Erickson AR, Pan C, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL (2013) Effects of diet on resource utilization by a model human gut microbiota containing bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol 11(8):e1001637
Murdoch JD, Buyandelger S, Cypher BL (2009) Patterns of seed occurrence in corsac and red fox diets in Mongolia. J Arid Environ 73:381–384
Delsuc F, Metcalf JL, Parfrey LW, Song SJ, Gonzalez A, Knight R (2014) Convergence of gut microbiome in myrmecophagous mammals. Mol Ecol 23:1301–1317
Whitaker JO, Dannelly HK, Prentice DA (2004) Chitinase in insectivorous bats. J Mammal 85:15–18
Hiscocks K, Perrin M (1987) Feeding observations and diet of black-backed jackals in an arid coastal environment. S Afr J Wildl Res 17:55–58
Axelsson E, Ratnakumar A, Arendt M-L, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar Å, Lindblad-Toh K (2013) The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495:360–364
Scott KP, Martin JC, Duncan SH, Flint HJ (2014) Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 87:30–40
Silverstein RL, Febbraio M (2009) CD36, a scavenger receptor involved in immunity, metabolism, angiogenesis, and behavior. Sci Signal 2(72):re3-re3
Hugenholtz F (2015) Mouse gut microbiomics of short chain fatty acid metabolism and mucosal responses. Doctoral dissertation, Wageningen University. https://library.wur.nl/WebQuery/wurpubs/482512
Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, Zhao J, Zhang H, Chen W (2021) Blautia—a new functional genus with potential probiotic properties? Gut Microbes 13:1–21
Jenq RR, Taur Y, Devlin SM, Ponce DM, Goldberg JD, Ahr KF, Littmann ER, Ling L, Gobourne AC, Miller LC (2015) Intestinal Blautia is associated with reduced death from graft-versus-host disease. Biol Blood Marrow Transplant 21:1373–1383
Xu S, Dong Y, Shi J, Li Z, Che L, Lin Y, Li J, Feng B, Fang Z, Yong Z, Wang J, Wu D (2021) Responses of vaginal microbiota to dietary supplementation with lysozyme and its relationship with rectal microbiota and sow performance from late gestation to early lactation. Animals 11:593
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systemS. Microbiol Mol Biol Rev 72:317–364
Qi C, Ding M, ** with fecal sample collection. We thank our mentors for their guidance in experiments and writing.
Funding
This work was supported by the Special Fund for Forest Scientific Research in the Public Welfare (201404420) and the National Natural Science Foundation of China (31872242, 32070405, 32001228, 32000291, 61902368).
Author information
Authors and Affiliations
Contributions
HhZ, XbW, and QgW conceived and designed the study. XbW, YqS, XyW, WlS, HxZ, and ScM performed the research. XbW, HsD, SyZ, and GlS analyzed the data. XbW prepared the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval (Include Appropriate Approvals or Waivers)
Applicable.
Consent to Participate (Include Appropriate Statements)
Applicable.
Consent for Publication (Include Appropriate Statements)
Applicable.
Competing Interests
The authors declare no competing interests.
Additional information
**bao Wang and Yongquan Shang shared first authorship
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Shang, Y., Wei, Q. et al. Comparative Analyses of the Gut Microbiome of Two Fox Species, the Red Fox (Vulpes Vulpes) and Corsac Fox (Vulpes Corsac), that Occupy Different Ecological Niches. Microb Ecol 83, 753–765 (2022). https://doi.org/10.1007/s00248-021-01806-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-021-01806-8