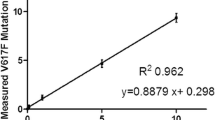
Abstract
Real-time quantitative PCR (qPCR) has been widely implemented for molecular testing, but there are still some inherent limitations that hamper its usefulness. Droplet digital PCR (ddPCR), which can provide direct, standards-free quantification, has recently received increasing attention. In our study, a comprehensive comparison of ddPCR with qPCR in relation to the quantification of PML-RARα was performed to evaluate the diagnostic potential of ddPCR. Results showed that ddPCR displayed significant concordance with qPCR in the detection of PML-RARα in clinical samples, but showed advantages over qPCR in terms of precision, limit of detection (LOD), and other basic performance parameters. A study of the feasibility of duplexing also indicated that ddPCR could simultaneously quantify the target PML-RARα and the clinical common reference gene ABL in a reaction, in contrast to qPCR. Moreover, ddPCR was more tolerant than qPCR of inhibition, and was shown to be able to quantify inhibition-prone samples. Another advantage of using ddPCR in clinical applications is that it will yield accurate results for patients with PML-RARα levels that fluctuate around the LOD of qPCR. Therefore, ddPCR is considered to have the potential to become a reliable alternative technique for quantifying PML-RARα.

ᅟ





Similar content being viewed by others
References
Cicconi L, Fenaux P, Kantarjian H, Tallman M, Sanz MA, Lo-Coco F. Molecular remission as a therapeutic objective in acute promyelocytic leukemia. Leukemia. 2018;32(8):1671–8.
Lo-Coco F, Ammatuna E. The biology of acute promyelocytic leukemia and its impact on diagnosis and treatment. Hematol Am Soc Hematol Educ Program. 2006;156-61:514.
Lo Coco F, Diverio D, Falini B, Biondi A, Nervi C, Pelicci PG. Genetic diagnosis and molecular monitoring in the management of acute promyelocytic leukemia. Blood. 1999;94(1):12–22.
Sanz MA, Lo-Coco F. Modern approaches to treating acute promyelocytic leukemia. J Clin Oncol. 2011;29(5):495–503.
Grimwade D, Jovanovic JV, Hills RK, Nugent EA, Patel Y, et al. Prospective minimal residual disease monitoring to predict relapse of acute promyelocytic leukemia and to direct pre-emptive arsenic trioxide therapy. J Clin Oncol. 2009;27(22):3650–8.
Bhat S, Emslie KR. Digital polymerase chain reaction for characterisation of DNA reference materials. Biomol Detect Quantif. 2016;10:47–9.
Waggoner J, Ho DY, Libiran P, Pinsky BA. Clinical significance of low cytomegalovirus DNA levels in human plasma. J Clin Microbiol. 2012;50(7):2378–83.
Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, et al. Evaluation of a droplet digital polymerase chain reaction format for DNA copy number quantification. Anal Chem. 2012;84(2):1003–11.
Huggett JF, Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem. 2013;59(12):1691–3.
Bizouarn F. Introduction to digital PCR. Methods Mol Biol. 2014;1160:27–41.
Albano F, Zagaria A, Anelli L, Coccaro N, Tota G, et al. Absolute quantification of the pretreatment PML-RARA transcript defines the relapse risk in acute promyelocytic leukemia. Oncotarget. 2015;6(15):13269–77.
Brunetti C, Anelli L, Zagaria A, Minervini A, Minervini CF, et al. Droplet digital PCR is a reliable tool for monitoring minimal residual disease in acute promyelocytic leukemia. J Mol Diagn. 2017;19(3):437–44.
He HJ, Almeida JL, Lund SP, Steffen CR, Choquette S, Cole KD. Development of NIST Standard Reference Material 2373: genomic DNA standards for HER2 measurements. Biomol Detect Quantif. 2016;8:1–8.
White H, Deprez L, Corbisier P, Hall V, Lin F, et al. A certified plasmid reference material for the standardisation of BCR-ABL1 mRNA quantification by real-time quantitative PCR. Leukemia. 2015;29(2):369–76.
Clinical and Laboratory Standards Institute, Tholen DW, Linnet K, Kondratovich M, Armbruster DA, Garrett PE, Jones RL, Kroll MH, Lequin RM, Pankratz TJ, Scassellati GA, Schimmel H, Tsai J. Protocols for determination of limits of detection and limits of quantitation. Approved guideline EP17-a. Wayne: Clinical and Laboratory Standards Institute; 2004.
Milosevic D, Mills JR, Campion MB, Vidal Folch N, Voss JS, et al. Applying standard clinical chemistry assay validation to droplet digital PCR quantitative liquid biopsy testing. Clin Chem. 2018.
Antonelli G, Padoan A, Aita A, Sciacovelli L, Plebani M. Verification of examination procedures in clinical laboratory for imprecision, trueness and diagnostic accuracy according to ISO 15189:2012: a pragmatic approach. Clin Chem Lab Med. 2017;55(10):1501–8.
Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin Chem. 2013;59(11):1670–2.
Bhat S, McLaughlin JL, Emslie KR. Effect of sustained elevated temperature prior to amplification on template copy number estimation using digital polymerase chain reaction. Analyst. 2011;136(4):724–32.
Cao L, Cui X, Hu J, Li Z, Choi JR, Yang Q, et al. Xu F. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron. 2017;90:459–74.
Cao Y, Raith MR, Griffith JF. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 2015;70:337–49.
Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61(1):79–88.
Sanders R, Huggett JF, Bushell CA, Cowen S, Scott DJ, Foy CA. Evaluation of digital PCR for absolute DNA quantification. Anal Chem. 2011;83(17):6474–84.
Morisset D, Stebih D, Milavec M, Gruden K, Zel J. Quantitative analysis of food and feed samples with droplet digital PCR. PLoS One. 2013;8(5):e62583.
Cao Y, Griffith JF, Dorevitch S, Weisberg SB. Effectiveness of qPCR permutations, internal controls and dilution as means for minimizing the impact of inhibition while measuring Enterococcus in environmental waters. J Appl Microbiol. 2012;113(1):66–75.
Schumacher JA, Scott Reading N, Szankasi P, Matynia AP, Kelley TW. A novel approach to quantitating leukemia fusion transcripts by qRT-PCR without the need for standard curves. Exp Mol Pathol. 2015;99(1):104–8.
Grimwade D, Jovanovic JV, Hills RK. Can we say farewell to monitoring minimal residual disease in acute promyelocytic leukaemia? Best Pract Res Clin Haematol. 2014;27(1):53–61.
Coccaro N, Anelli L, Zagaria A, Casieri P, Tota G, et al. Droplet digital PCR is a robust tool for monitoring minimal residual disease in adult Philadelphia-positive acute lymphoblastic leukemia. J Mol Diagn. 2018;20(4):474–82.
Verhaegen B, De Reu K, De Zutter L, Verstraete K, Heyndrickx M, Van Coillie E. Comparison of droplet digital PCR and qPCR for the quantification of Shiga toxin-producing Escherichia coli in bovine feces. Toxins (Basel) 2016;8(5).
Jones GM, Busby E, Garson JA, Grant PR, Nastouli E, Devonshire AS, et al. Digital PCR dynamic range is approaching that of real-time quantitative PCR. Biomol Detect Quantif. 2016;10:31–3.
Vynck M, Trypsteen W, Thas O, Vandekerckhove L, De Spiegelaere W. The future of digital polymerase chain reaction in virology. Mol Diagn Ther. 2016;20(5):437–47.
Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, et al. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59(6):892–902.
Kuypers J, Jerome KR. Applications of digital PCR for clinical microbiology. J Clin Microbiol. 2017;55(6):1621–8.
Hall Sedlak R, Jerome KR. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev Mol Diagn. 2014;14(4):501–7.
Basu AS. Digital assays part I: partitioning statistics and digital PCR. SLAS Technol. 2017;22(4):369–86.
Song Q, Gao Y, Zhu Q, Tian Q, Yu B, et al. A nanoliter self-priming compartmentalization chip for point-of-care digital PCR analysis. Biomed Microdevices. 2015;17(3):64.
Sinha M, Mack H, Coleman TP, Fraley SI. A high-resolution digital DNA melting platform for robust sequence profiling and enhanced genotype discrimination. SLAS Technol. 2018;23(6):580–91.
Acknowledgements
This study was supported by the National Natural Science Foundation (grant numbers 81201349, 81000775); Young Medical Key Talents in Jiangsu Province (grant numbers QNRC 2016686, 2016687); Frontier and Key Technical Innovation Projects of Nantong (grant number MS22015049); and the Nantong Science and Technology Plan Project (MS12017008-3).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
The authors declare that all individual participants from whom the blood samples were obtained gave their informed consent, and that the studies were approved by the ethics committee of the Affiliated Hospital of Nantong University and were performed in accordance with ethical standards.
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 322 kb)
Rights and permissions
About this article
Cite this article
Yuan, D., Cui, M., Yu, S. et al. Droplet digital PCR for quantification of PML-RARα in acute promyelocytic leukemia: a comprehensive comparison with real-time PCR. Anal Bioanal Chem 411, 895–903 (2019). https://doi.org/10.1007/s00216-018-1508-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-018-1508-6